Trick or tips 004 {R}
August 13, 2019
R tips trickortips

Table of Contents
Ever tumbled on a code chunk that made you say: "I should have known this f_ piece of code long ago!" Chances are you have, frustratingly, just like we have, and on multiple occasions too. In comes Trick or Tips!
Trick or Tips is a series of blog posts that each present 5 -- hopefully helpful -- coding tips for a specific programming language. Posts should be short (i.e. no more than 5 lines of code, max 80 characters per line, except when appropriate) and provide tips of many kind: a function, a way of combining of functions, a single argument, a note about the philosophy of the language and practical consequences, tricks to improve the way you code, good practices, etc.
Note that while some tips might be obvious for careful documentation readers (God bless them for their wisdom), we do our best to present what we find very useful and underestimated. By the way, there are undoubtedly similar initiatives on the web (e.g. "One R Tip a Day" Twitter account). Last, feel free to comment below tip ideas or a post of code tips of your own which we will be happy to incorporate to our next post.
Enjoy and get ready to frustratingly appreciate our tips!
Subset an array with a matrix
Let’s consider two arrays of letters: the first has two dimensions (i.e. a matrix) and the second one has 3.
|
|
Let’s say, you need to subset a specific set of values based on the position of the elements. To subset a single element, say "G", there are a couple of options, but I guess the most common approach is to use [ with one value per dimension:
|
|
or with a single value giving the position of the element:
|
|
Now we consider the case where you have a vector of positions (one value per dimension of the array), in this case, beware the orientation of the vector!
|
|
And for more than one element, you need to use a matrix with one row per element to be subset:
|
|
Similarly, with an array of 3 dimensions, the matrix will have three columns and as many row as there are elements:
|
|
Two additional comments. First, we should always keep in mind that data frames and arrays are different:
|
|
Second, if you are a tidyverse user, there is a new article dealing with subassigment with tibble 😎.
nzchar()
You may already be aware of nchar(), a function that returns the number of characters of a given character vector:
|
|
nzchar() returns TRUE for every character string in the vector that has at least 1 character:
|
|
Interesting, but let’s dig deeper: I can think about no less than 3 ways of writing a equivalent function with one more character:
|
|
One more character… so why bother? 💡 It should be a matter of performance! Let’s check that out with the cool 📦 microbenchmark:
|
|
Yep yep!nzchar() is indeed way faster 🚀!
Do you need to use return()?
If you have already written your own function, you must have used return()
to specify what your function should return. There are programming languages where this instruction is mandatory, not in R! Check out the documentation ?return:
If the end of a function is reached without calling ‘return’, the value of the last evaluated expression is returned.
Let me write 2 functions:
|
|
add_v() and add_v2() are equivalent! So… do we care? Well, you must bear in mind that whenever return() is encountered, the evaluation of the set of expressions within the function is stopped and therefore some time can be saved:
|
|
invisible()
Let’s keep talking about what functions return. The function invisible() allows you to return an invisible copy of an object, meaning that nothing is (apparently) return if not assigned:
|
|
But… why? As explained in the documentation (?invisible):
This function can be useful when it is desired to have functions return values which can be assigned, but which do not print when they are not assigned.
This is indeed helpful when you have a function that creates a plot (and you don’t normally to assign the result) for which you sometimes need to use an object that was created during the evaluation of the function:
|
|
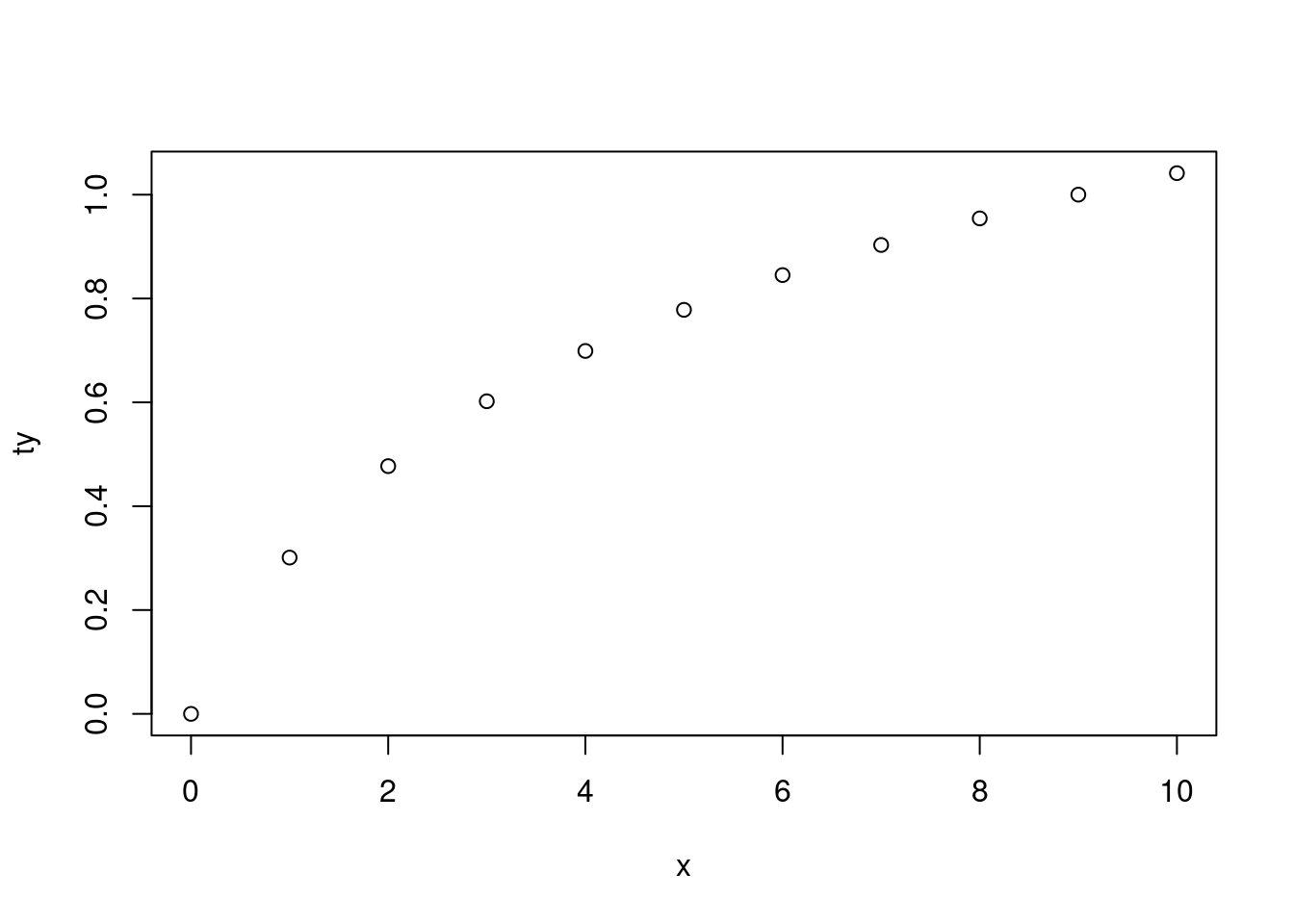
|
|
|
|
bquote() and substitute()
When using mathematical annotations, we sometimes need to include the value
of a variable. In such case, bquote() or substitute() are the functions you
would need (rather than expression() you may already be familiar with).
If you opt for bquote(), then variables to be evaluated must be put in
brackets and preceded by a dot, e.g. .(var). If you choose substitute(),
then variables evaluated will be the ones included in the list passed as
argument env (which can also be the name of a environment).
Let’s use both functions in to add mathematical expressions in an empty plot:
|
|
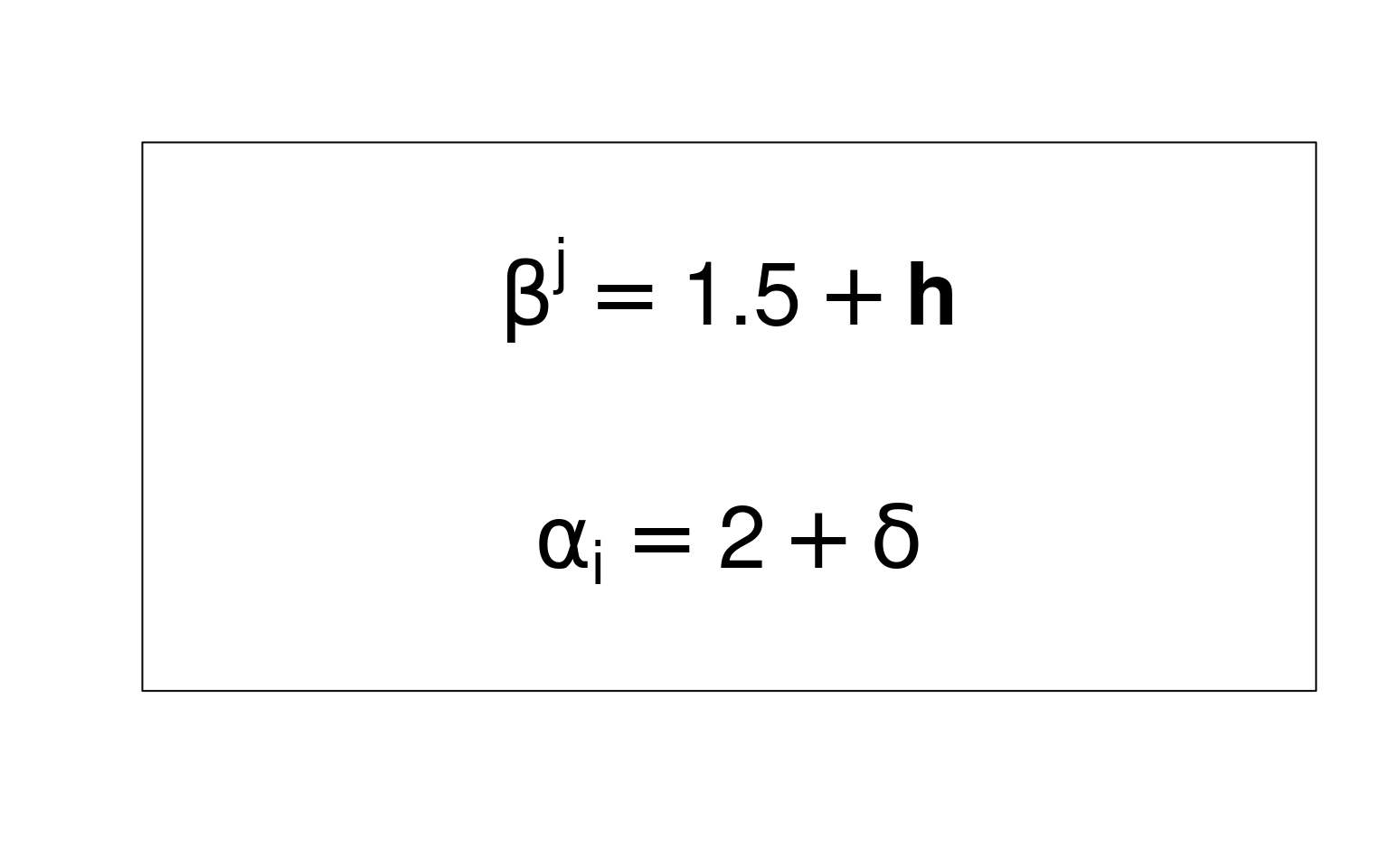
|
|
That’s all folks!
Display information relative to the R session used to render this post.
|
|
Edits
Feb 6, 2023 -- Remove redundant headers.



