<br><br><br> .maintitle[<i class="fa fa-angle-double-right" aria-hidden="true"></i>intRoduction] <hr> <br> ### Marie-Hélène Brice ### Kevin Cazelles #### UQAM - March 26th, 2019 -- [<i class="fa fa-github" aria-hidden="true"></i>](https://github.com/inSileco/IntroR) .right[   ] --- # Aims <br> ### Better ideas about what R actually is ### Better ideas about what R can do ### Better ideas about the topics covered --- # Outline ### <i class="fa fa-commenting-o" aria-hidden="true"></i> R? ~10min ### <i class="fa fa-wrench" aria-hidden="true"></i> Data manipulations ~1h15min ### <i class="fa fa-bar-chart" aria-hidden="true"></i> Data visualization (part1) ~30min ### <i class="fa fa-cutlery" aria-hidden="true"></i> Lunch break 12pm ### <i class="fa fa-bar-chart" aria-hidden="true"></i> Data visualization (part2) ~30min ### <i class="fa fa-map" aria-hidden="true"></i> mapping ~ 1h30 ### <i class="fa fa-coffee" aria-hidden="true"></i> *coffee break* 3pm ### <i class="fa fa-sitemap" aria-hidden="true"></i> ordination ~1h30 --- class: inverse, center, middle # R? ## <i class="fa fa-commenting-o" aria-hidden="true"></i> --- # R <br> > R is a programming language and free software environment for statistical computing and graphics [...]. <br> > The R language is widely used among statisticians and data miners for developing statistical software and data analysis. [R (programming language) - Wikipedia](https://en.wikipedia.org/wiki/R_(programming_language) --- # R is popular .center[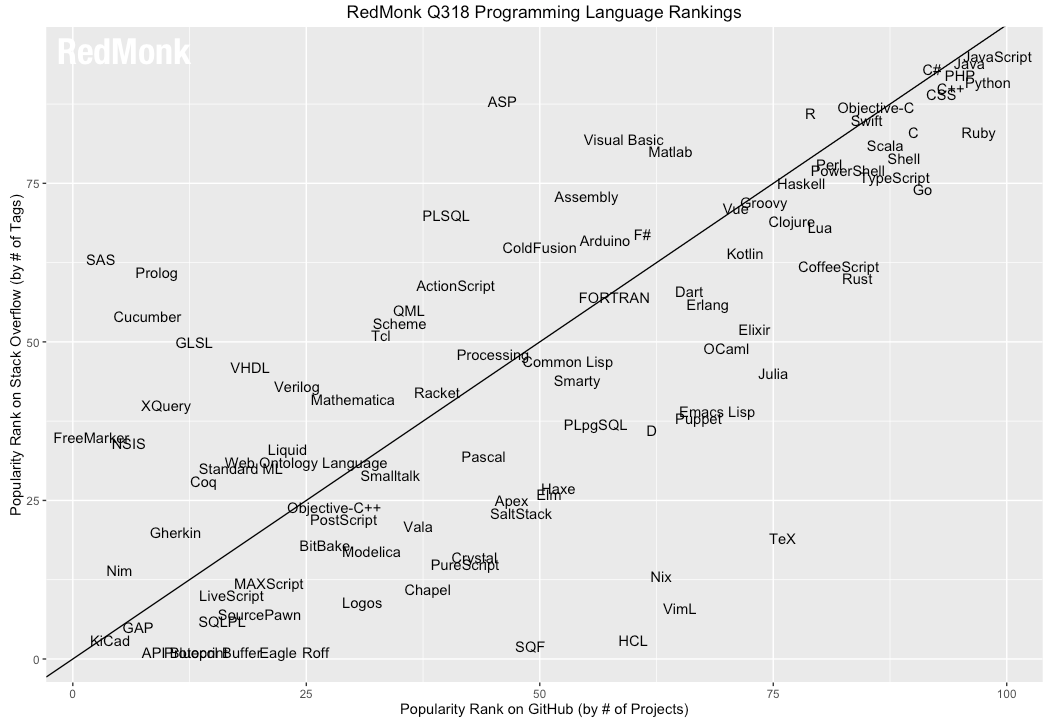] [Python](https://www.python.org/) / [Julia](https://julialang.org/) --- # R is popular among Biological Sciences ### For instance 1. Ecologists 2. Bioinformatics [(Bioconductor)](https://www.bioconductor.org/) 3. Medicine [<i class="fa fa-external-link" aria-hidden="true"></i>](https://r-medicine.com/) -- ### Why? 1. An incredible tool for statistics 2. Fairly accessible for non-programmer people 3. A very high diversity of packages 4. Improved workflow --- # R is popular .center[] [*CRAN now has 10,000 R packages* Revolutions. January, 2017](https://blog.revolutionanalytics.com/2017/01/cran-10000.html) [MetaCRAN](https://www.r-pkg.org/) / [R Package Documentation](https://rdrr.io/) --- # R, RStudio, Ropenscience? ### Main links - [R project](https://www.r-project.org/) - [CRAN](https://cran.r-project.org/) - [RStudio](https://www.rstudio.com/) - [rOpenSci](https://ropensci.org) ### Get reliable documentation - [CRAN Manual & Contributed](https://cran.r-project.org/) - [bookdown](https://bookdown.org/) - [RStudio](https://www.rstudio.com/) - [Cheat Sheets](https://www.rstudio.com/resources/cheatsheets/) - [Webinars](https://resources.rstudio.com/webinars) - [QCBS](https://qcbsrworkshops.github.io/Workshops/) - [DataCarpentery](https://datacarpentry.org/R-genomics/index.html) - [<i class="fa fa-stack-overflow" aria-hidden="true"></i>](https://stackoverflow.com/questions/tagged/r) --- class: inverse, center, middle # Data manipulation ## <i class="fa fa-wrench" aria-hidden="true"></i> --- # Let's start <br> - Open R console / R GUI / RStudio / ... -- ```R getwd() setwd("pasth2workingdirectory") ``` -- - Install the following packages ```R install.packages(c("tidyverse", "sf", "raster", "mapview", "vegan", "ade4", "scales", "Rcolorbrewer")) ``` --- # R's principles <br> ### 1. Everything in R is an object ### 2. Everything that happens in R is a function call ### 3. Interfaces to other software are part of R. [Extending-R by Chambers](https://www.crcpress.com/Extending-R/Chambers/p/book/9781498775717) --- # Everything in R is an object <br> ```r 2 # [1] 2 ``` -- ```r class(2) # [1] "numeric" ``` -- ```r class("A") # [1] "character" class(library) # [1] "function" plot # function (x, y, ...) # UseMethod("plot") # <bytecode: 0x56490cfc97f8> # <environment: namespace:graphics> ``` --- class: center # Everything that happens in R is a function call <br> ```R class(2) ``` ## <i class="fa fa-arrow-circle-o-down" aria-hidden="true"></i> ### `enter` ## <i class="fa fa-arrow-circle-o-down" aria-hidden="true"></i> ``` # [1] "numeric" ``` --- # Interfaces to other software are part of R <br> ### Four examples among many others: - [Rcpp](https://cran.r-project.org/web/packages/Rcpp/index.html) - [Reticulate](https://rstudio.github.io/reticulate/index.html) - [Rmarkdown](https://rmarkdown.rstudio.com/) - [plotly](https://plot.ly/r/) - [mapview](https://r-spatial.github.io/mapview/) --- # Basic commands ```r 2+2 # [1] 4 2*3 # [1] 6 3>2 # [1] TRUE ``` -- ```r let <- LETTERS[1:10] let2 = LETTERS[1:10] let # [1] "A" "B" "C" "D" "E" "F" "G" "H" "I" "J" let2 # [1] "A" "B" "C" "D" "E" "F" "G" "H" "I" "J" identical(let, let2) # [1] TRUE ``` -- ```r let3 <- sample(let, 5) let3 # [1] "J" "G" "H" "I" "E" ``` -- ```R # Functions documentation ?sample ``` --- # Vectors **Create a vector** ```r vec <- c(2:5, 9:12, 2) vec # [1] 2 3 4 5 9 10 11 12 2 vec > 4 # [1] FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE FALSE ``` -- ```r let # [1] "A" "B" "C" "D" "E" "F" "G" "H" "I" "J" length(let) # [1] 10 ``` -- **Subset a vector** ```r let[1:2] # [1] "A" "B" let[c(1:2, 5:6)] # [1] "A" "B" "E" "F" let[-c(1,7)] # [1] "B" "C" "D" "E" "F" "H" "I" "J" vec[vec > 4] # [1] 5 9 10 11 12 let[let > "C"] # [1] "D" "E" "F" "G" "H" "I" "J" ``` --- # Matrices ```r # use arguments mat <- matrix(1:18, ncol = 3, nrow = 6) mat # [,1] [,2] [,3] # [1,] 1 7 13 # [2,] 2 8 14 # [3,] 3 9 15 # [4,] 4 10 16 # [5,] 5 11 17 # [6,] 6 12 18 dim(mat) # [1] 6 3 mat[1,2] # [1] 7 ``` -- ```r mat[1,] # [1] 1 7 13 mat[,2] # [1] 7 8 9 10 11 12 mat[2:4, 1:2] # [,1] [,2] # [1,] 2 8 # [2,] 3 9 # [3,] 4 10 ``` --- # Data frames ## <i class="fa fa-question-circle-o" aria-hidden="true"></i> -- ```r df <- data.frame( letter = let[1:5], val = vec[1:5], logic = vec[1:5]>2 ) df # letter val logic # 1 A 2 FALSE # 2 B 3 TRUE # 3 C 4 TRUE # 4 D 5 TRUE # 5 E 9 TRUE class(df) # [1] "data.frame" dim(df) # [1] 5 3 ``` --- # Data frames ```r df[1, 1] # [1] "A" ``` ```r df$letter # [1] "A" "B" "C" "D" "E" df[,1] # [1] "A" "B" "C" "D" "E" df[1] # letter # 1 A # 2 B # 3 C # 4 D # 5 E df[1:2] # letter val # 1 A 2 # 2 B 3 # 3 C 4 # 4 D 5 # 5 E 9 ``` ```r class(df$letter) # [1] "character" class(df$val) # [1] "numeric" ``` --- # Data frames <br> ```r library(datasets) ``` To see the list of available datasets: ```R data() ``` To access the documentation of a particular dataset: ```R ?CO2 ``` --- # Data frames ```r library(datasets) head(CO2) # Grouped Data: uptake ~ conc | Plant # Plant Type Treatment conc uptake # 1 Qn1 Quebec nonchilled 95 16.0 # 2 Qn1 Quebec nonchilled 175 30.4 # 3 Qn1 Quebec nonchilled 250 34.8 # 4 Qn1 Quebec nonchilled 350 37.2 # 5 Qn1 Quebec nonchilled 500 35.3 # 6 Qn1 Quebec nonchilled 675 39.2 names(CO2) # [1] "Plant" "Type" "Treatment" "conc" "uptake" summary(CO2) # Plant Type Treatment conc uptake # Qn1 : 7 Quebec :42 nonchilled:42 Min. : 95 Min. : 7.70 # Qn2 : 7 Mississippi:42 chilled :42 1st Qu.: 175 1st Qu.:17.90 # Qn3 : 7 Median : 350 Median :28.30 # Qc1 : 7 Mean : 435 Mean :27.21 # Qc3 : 7 3rd Qu.: 675 3rd Qu.:37.12 # Qc2 : 7 Max. :1000 Max. :45.50 # (Other):42 ``` -- ```r CO2$Plant # [1] Qn1 Qn1 Qn1 Qn1 Qn1 Qn1 Qn1 Qn2 Qn2 Qn2 Qn2 Qn2 Qn2 Qn2 Qn3 Qn3 Qn3 Qn3 Qn3 Qn3 Qn3 Qc1 # [23] Qc1 Qc1 Qc1 Qc1 Qc1 Qc1 Qc2 Qc2 Qc2 Qc2 Qc2 Qc2 Qc2 Qc3 Qc3 Qc3 Qc3 Qc3 Qc3 Qc3 Mn1 Mn1 # [45] Mn1 Mn1 Mn1 Mn1 Mn1 Mn2 Mn2 Mn2 Mn2 Mn2 Mn2 Mn2 Mn3 Mn3 Mn3 Mn3 Mn3 Mn3 Mn3 Mc1 Mc1 Mc1 # [67] Mc1 Mc1 Mc1 Mc1 Mc2 Mc2 Mc2 Mc2 Mc2 Mc2 Mc2 Mc3 Mc3 Mc3 Mc3 Mc3 Mc3 Mc3 # Levels: Qn1 < Qn2 < Qn3 < Qc1 < Qc3 < Qc2 < Mn3 < Mn2 < Mn1 < Mc2 < Mc3 < Mc1 ``` --- # Lists ```r mylist <- list(CO2 = CO2, mymat = mat, mylet = let[1], awesome = "cool") names(mylist) # [1] "CO2" "mymat" "mylet" "awesome" mylist$awesome # [1] "cool" mylist[3:4] # $mylet # [1] "A" # # $awesome # [1] "cool" ``` -- ```r class(mylist[2]) # [1] "list" class(mylist[[2]]) # [1] "matrix" mylist[[2]] # [,1] [,2] [,3] # [1,] 1 7 13 # [2,] 2 8 14 # [3,] 3 9 15 # [4,] 4 10 16 # [5,] 5 11 17 # [6,] 6 12 18 class(mylist[[2]][2,3]) # [1] "integer" mylist[[2]][2,3] # [1] 14 ``` --- # For loops ```r for (i in 1:4) { # actions print(i) } # [1] 1 # [1] 2 # [1] 3 # [1] 4 ``` ```r for (j in LETTERS[1:5]) { print(j) } # [1] "A" # [1] "B" # [1] "C" # [1] "D" # [1] "E" ``` ```r for (i in c(1,4,6)) { print(i) } # [1] 1 # [1] 4 # [1] 6 ``` --- # Logical conditions > >= < <= == != -- ```r vec <- c(2:5, 9:12, 2) vec # [1] 2 3 4 5 9 10 11 12 2 vec[1] > 4 # [1] FALSE vec > 4 # [1] FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE FALSE ``` -- `if` ... `else` ... ```r for (i in vec) { if (i <= 5) { # actions print(i) } else { print("nope") } } # [1] 2 # [1] 3 # [1] 4 # [1] 5 # [1] "nope" # [1] "nope" # [1] "nope" # [1] "nope" # [1] 2 ``` --- class: inverse, center, middle # Questions? ## <i class="fa fa-question-circle-o" aria-hidden="true"></i> --- # Tidyverse - a metapackage <br> ```r library(tidyverse) ``` ### <i class="fa fa-wrench" aria-hidden="true"></i> A great tool belt for data science [<i class="fa fa-external-link" aria-hidden="true"></i>](https://www.tidyverse.org/) -- - <i class="fa fa-check" aria-hidden="true"></i> Pros: - well-documented, intuitive - efficient - very popular - <i class="fa fa-exclamation-triangle" aria-hidden="true"></i>Cons: - an alternative way of doing the same manipulation things - prevents the user from learning programming basics --- # Data manipulations <br> ### 1. Import data / read file(s) <i class="fa fa-arrow-circle-right" aria-hidden="true"></i> get R object(s) ### 2. Select / Filter <i class="fa fa-arrow-circle-right" aria-hidden="true"></i> find the data of interest ### 3. Join <i class="fa fa-arrow-circle-right" aria-hidden="true"></i> combine data ### 4. Mutate / Aggregate <i class="fa fa-arrow-circle-right" aria-hidden="true"></i> create new data ### 5. Cast and Melt <i class="fa fa-arrow-circle-right" aria-hidden="true"></i> Format your data (see tidyr) ### 6. R object(s) <i class="fa fa-arrow-circle-right" aria-hidden="true"></i> Export data / write file(s) --- # Read a file ```r df2 <- read.csv("data/environ.csv") head(df2, 3) # dfs alt slo flo pH har pho nit amm oxy bdo # 1 3 934 6.176 84 79 45 1 20 0 122 27 # 2 22 932 3.434 100 80 40 2 20 10 103 19 # 3 102 914 3.638 180 83 52 5 22 5 105 35 class(df2) # [1] "data.frame" ``` -- ```r df3 <- read_csv("data/environ.csv") head(df3, 3) # # A tibble: 3 x 11 # dfs alt slo flo pH har pho nit amm oxy bdo # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 3 934 6.18 84 79 45 1 20 0 122 27 # 2 22 932 3.43 100 80 40 2 20 10 103 19 # 3 102 914 3.64 180 83 52 5 22 5 105 35 class(df3) # [1] "spec_tbl_df" "tbl_df" "tbl" "data.frame" ``` -- - `read.table()` - [SQL in R](http://dept.stat.lsa.umich.edu/~jerrick/courses/stat701/notes/sql.html) - [MongoDB in R](https://jeroen.github.io/mongolite/) - [MariaDB](https://mariadb.com/kb/en/library/r-statistical-programming-using-mariadb-as-the-background-database/) --- # Doubs River Fish Dataset <br> .pull-left[ Verneaux (1973) dataset: - characterization of fish communities - 27 different species - 30 different sites - 11 environmental variables Load the Doubs River species data ```r library(ade4) data(doubs) # ?doubs ``` ] .pull.right[ 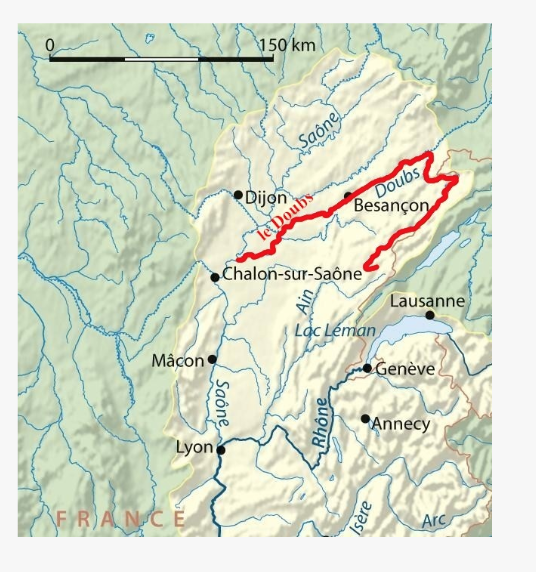 ] --- # Dataset 'doubs' in package 'ade4' <br> ```r class(doubs) # [1] "list" names(doubs) # [1] "env" "fish" "xy" "species" head(doubs$env) # dfs alt slo flo pH har pho nit amm oxy bdo # 1 3 934 6.176 84 79 45 1 20 0 122 27 # 2 22 932 3.434 100 80 40 2 20 10 103 19 # 3 102 914 3.638 180 83 52 5 22 5 105 35 # 4 185 854 3.497 253 80 72 10 21 0 110 13 # 5 215 849 3.178 264 81 84 38 52 20 80 62 # 6 324 846 3.497 286 79 60 20 15 0 102 53 ``` --- # Dataset 'doubs' in package 'ade4' <br> ```r as_tibble(doubs$env) # # A tibble: 30 x 11 # dfs alt slo flo pH har pho nit amm oxy bdo # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 3 934 6.18 84 79 45 1 20 0 122 27 # 2 22 932 3.43 100 80 40 2 20 10 103 19 # 3 102 914 3.64 180 83 52 5 22 5 105 35 # 4 185 854 3.50 253 80 72 10 21 0 110 13 # 5 215 849 3.18 264 81 84 38 52 20 80 62 # 6 324 846 3.50 286 79 60 20 15 0 102 53 # 7 268 841 4.20 400 81 88 7 15 0 111 22 # 8 491 792 3.26 130 81 94 20 41 12 70 81 # 9 705 752 2.56 480 80 90 30 82 12 72 52 # 10 990 617 4.61 1000 77 82 6 75 1 100 43 # # … with 20 more rows ``` --- # Piping -- ```r res <- as_tibble(data.frame(res = log(diff(exp(1:6))))) res # # A tibble: 5 x 1 # res # <dbl> # 1 1.54 # 2 2.54 # 3 3.54 # 4 4.54 # 5 5.54 ``` -- ```r # library(magrittr) res <- 1:6 %>% exp %>% diff %>% log %>% as.data.frame(col.names = "res") %>% as_tibble res # # A tibble: 5 x 1 # . # <dbl> # 1 1.54 # 2 2.54 # 3 3.54 # 4 4.54 # 5 5.54 ``` --- # Select -- ```r denv <- as_tibble(doubs$env) dim(denv) # [1] 30 11 names(denv) # [1] "dfs" "alt" "slo" "flo" "pH" "har" "pho" "nit" "amm" "oxy" "bdo" head(denv, 20) # # A tibble: 20 x 11 # dfs alt slo flo pH har pho nit amm oxy bdo # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 3 934 6.18 84 79 45 1 20 0 122 27 # 2 22 932 3.43 100 80 40 2 20 10 103 19 # 3 102 914 3.64 180 83 52 5 22 5 105 35 # 4 185 854 3.50 253 80 72 10 21 0 110 13 # 5 215 849 3.18 264 81 84 38 52 20 80 62 # 6 324 846 3.50 286 79 60 20 15 0 102 53 # 7 268 841 4.20 400 81 88 7 15 0 111 22 # 8 491 792 3.26 130 81 94 20 41 12 70 81 # 9 705 752 2.56 480 80 90 30 82 12 72 52 # 10 990 617 4.61 1000 77 82 6 75 1 100 43 # 11 1234 483 3.74 1990 81 96 30 160 0 115 27 # 12 1324 477 2.83 2000 79 86 4 50 0 122 30 # 13 1436 450 3.09 2110 81 98 6 52 0 124 24 # 14 1522 434 2.56 2120 83 98 27 123 0 123 38 # 15 1645 415 1.79 2300 86 86 40 100 0 117 21 # 16 1859 375 3.04 1610 80 88 20 200 5 103 27 # 17 1985 348 1.79 2430 80 92 20 250 20 102 46 # 18 2110 332 2.20 2500 80 90 50 220 20 103 28 # 19 2246 310 1.79 2590 81 84 60 220 15 106 33 # 20 2477 286 2.20 2680 80 86 30 300 30 103 28 ``` --- # Select ```r head(denv[, c(1:2, 5)]) # # A tibble: 6 x 3 # dfs alt pH # <dbl> <dbl> <dbl> # 1 3 934 79 # 2 22 932 80 # 3 102 914 83 # 4 185 854 80 # 5 215 849 81 # 6 324 846 79 head(denv[, c("dfs", "alt", "pH")]) # # A tibble: 6 x 3 # dfs alt pH # <dbl> <dbl> <dbl> # 1 3 934 79 # 2 22 932 80 # 3 102 914 83 # 4 185 854 80 # 5 215 849 81 # 6 324 846 79 ``` --- # Select ```r denvS <- denv %>% select(dfs, alt, pH) head(denvS) # # A tibble: 6 x 3 # dfs alt pH # <dbl> <dbl> <dbl> # 1 3 934 79 # 2 22 932 80 # 3 102 914 83 # 4 185 854 80 # 5 215 849 81 # 6 324 846 79 ``` ```r denvS <- denv %>% select(alt, pH, dfs) head(denvS) # # A tibble: 6 x 3 # alt pH dfs # <dbl> <dbl> <dbl> # 1 934 79 3 # 2 932 80 22 # 3 914 83 102 # 4 854 80 185 # 5 849 81 215 # 6 846 79 324 ``` --- # Select ```r head(denv[, -c(1:2, 5)]) # # A tibble: 6 x 8 # slo flo har pho nit amm oxy bdo # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 6.18 84 45 1 20 0 122 27 # 2 3.43 100 40 2 20 10 103 19 # 3 3.64 180 52 5 22 5 105 35 # 4 3.50 253 72 10 21 0 110 13 # 5 3.18 264 84 38 52 20 80 62 # 6 3.50 286 60 20 15 0 102 53 # head(denv[, -c("dfs", "alt", "pH")]) won't work ``` ```r denv %>% select(-dfs, -alt, -pH) # # A tibble: 30 x 8 # slo flo har pho nit amm oxy bdo # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 6.18 84 45 1 20 0 122 27 # 2 3.43 100 40 2 20 10 103 19 # 3 3.64 180 52 5 22 5 105 35 # 4 3.50 253 72 10 21 0 110 13 # 5 3.18 264 84 38 52 20 80 62 # 6 3.50 286 60 20 15 0 102 53 # 7 4.20 400 88 7 15 0 111 22 # 8 3.26 130 94 20 41 12 70 81 # 9 2.56 480 90 30 82 12 72 52 # 10 4.61 1000 82 6 75 1 100 43 # # … with 20 more rows ``` --- # Filter ```r denv$alt # [1] 934 932 914 854 849 846 841 792 752 617 483 477 450 434 415 375 348 332 310 286 262 254 # [23] 246 241 231 214 206 195 183 172 ``` -- ```r denvF <- denv %>% dplyr::filter(alt > 400) denvF # # A tibble: 15 x 11 # dfs alt slo flo pH har pho nit amm oxy bdo # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 3 934 6.18 84 79 45 1 20 0 122 27 # 2 22 932 3.43 100 80 40 2 20 10 103 19 # 3 102 914 3.64 180 83 52 5 22 5 105 35 # 4 185 854 3.50 253 80 72 10 21 0 110 13 # 5 215 849 3.18 264 81 84 38 52 20 80 62 # 6 324 846 3.50 286 79 60 20 15 0 102 53 # 7 268 841 4.20 400 81 88 7 15 0 111 22 # 8 491 792 3.26 130 81 94 20 41 12 70 81 # 9 705 752 2.56 480 80 90 30 82 12 72 52 # 10 990 617 4.61 1000 77 82 6 75 1 100 43 # 11 1234 483 3.74 1990 81 96 30 160 0 115 27 # 12 1324 477 2.83 2000 79 86 4 50 0 122 30 # 13 1436 450 3.09 2110 81 98 6 52 0 124 24 # 14 1522 434 2.56 2120 83 98 27 123 0 123 38 # 15 1645 415 1.79 2300 86 86 40 100 0 117 21 ``` --- # Filter <br> ```r denvF2 <- denv %>% dplyr::filter(alt > 400 & pH>=80) denvF2 # # A tibble: 11 x 11 # dfs alt slo flo pH har pho nit amm oxy bdo # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 22 932 3.43 100 80 40 2 20 10 103 19 # 2 102 914 3.64 180 83 52 5 22 5 105 35 # 3 185 854 3.50 253 80 72 10 21 0 110 13 # 4 215 849 3.18 264 81 84 38 52 20 80 62 # 5 268 841 4.20 400 81 88 7 15 0 111 22 # 6 491 792 3.26 130 81 94 20 41 12 70 81 # 7 705 752 2.56 480 80 90 30 82 12 72 52 # 8 1234 483 3.74 1990 81 96 30 160 0 115 27 # 9 1436 450 3.09 2110 81 98 6 52 0 124 24 # 10 1522 434 2.56 2120 83 98 27 123 0 123 38 # 11 1645 415 1.79 2300 86 86 40 100 0 117 21 ``` --- # Filter <br> ```r denvF3 <- denv %>% dplyr::filter(alt > 600 | pH>=82) denvF3 # # A tibble: 14 x 11 # dfs alt slo flo pH har pho nit amm oxy bdo # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 3 934 6.18 84 79 45 1 20 0 122 27 # 2 22 932 3.43 100 80 40 2 20 10 103 19 # 3 102 914 3.64 180 83 52 5 22 5 105 35 # 4 185 854 3.50 253 80 72 10 21 0 110 13 # 5 215 849 3.18 264 81 84 38 52 20 80 62 # 6 324 846 3.50 286 79 60 20 15 0 102 53 # 7 268 841 4.20 400 81 88 7 15 0 111 22 # 8 491 792 3.26 130 81 94 20 41 12 70 81 # 9 705 752 2.56 480 80 90 30 82 12 72 52 # 10 990 617 4.61 1000 77 82 6 75 1 100 43 # 11 1522 434 2.56 2120 83 98 27 123 0 123 38 # 12 1645 415 1.79 2300 86 86 40 100 0 117 21 # 13 3947 195 1.39 4320 83 100 74 400 30 81 45 # 14 4530 172 1.10 6900 82 109 65 160 10 82 44 ``` --- # Filter <br> Selection + Filter -- ```r denvF4 <- denv %>% select(dfs, alt, pH) %>% dplyr::filter(alt > 400 & pH>=80) denvF4 # # A tibble: 11 x 3 # dfs alt pH # <dbl> <dbl> <dbl> # 1 22 932 80 # 2 102 914 83 # 3 185 854 80 # 4 215 849 81 # 5 268 841 81 # 6 491 792 81 # 7 705 752 80 # 8 1234 483 81 # 9 1436 450 81 # 10 1522 434 83 # 11 1645 415 86 ``` --- # Mutate <br> -- ```r denvM <- denv %>% mutate(pH2 = pH + 1) denvM # # A tibble: 30 x 12 # dfs alt slo flo pH har pho nit amm oxy bdo pH2 # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 3 934 6.18 84 79 45 1 20 0 122 27 80 # 2 22 932 3.43 100 80 40 2 20 10 103 19 81 # 3 102 914 3.64 180 83 52 5 22 5 105 35 84 # 4 185 854 3.50 253 80 72 10 21 0 110 13 81 # 5 215 849 3.18 264 81 84 38 52 20 80 62 82 # 6 324 846 3.50 286 79 60 20 15 0 102 53 80 # 7 268 841 4.20 400 81 88 7 15 0 111 22 82 # 8 491 792 3.26 130 81 94 20 41 12 70 81 82 # 9 705 752 2.56 480 80 90 30 82 12 72 52 81 # 10 990 617 4.61 1000 77 82 6 75 1 100 43 78 # # … with 20 more rows ``` --- # Mutate <br> ```r denvM2 <- denv %>% mutate(index = 2*nit + pho + amm) denvM2 # # A tibble: 30 x 12 # dfs alt slo flo pH har pho nit amm oxy bdo index # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 3 934 6.18 84 79 45 1 20 0 122 27 41 # 2 22 932 3.43 100 80 40 2 20 10 103 19 52 # 3 102 914 3.64 180 83 52 5 22 5 105 35 54 # 4 185 854 3.50 253 80 72 10 21 0 110 13 52 # 5 215 849 3.18 264 81 84 38 52 20 80 62 162 # 6 324 846 3.50 286 79 60 20 15 0 102 53 50 # 7 268 841 4.20 400 81 88 7 15 0 111 22 37 # 8 491 792 3.26 130 81 94 20 41 12 70 81 114 # 9 705 752 2.56 480 80 90 30 82 12 72 52 206 # 10 990 617 4.61 1000 77 82 6 75 1 100 43 157 # # … with 20 more rows ``` --- # Mutate ```r denvM3 <- denv %>% select(dfs, alt, pH, nit, pho, amm) %>% dplyr::filter(alt > 400) %>% mutate(pH2 = pH + 1) %>% mutate(index = 2*nit + pho + amm) denvM3 # # A tibble: 15 x 8 # dfs alt pH nit pho amm pH2 index # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 3 934 79 20 1 0 80 41 # 2 22 932 80 20 2 10 81 52 # 3 102 914 83 22 5 5 84 54 # 4 185 854 80 21 10 0 81 52 # 5 215 849 81 52 38 20 82 162 # 6 324 846 79 15 20 0 80 50 # 7 268 841 81 15 7 0 82 37 # 8 491 792 81 41 20 12 82 114 # 9 705 752 80 82 30 12 81 206 # 10 990 617 77 75 6 1 78 157 # 11 1234 483 81 160 30 0 82 350 # 12 1324 477 79 50 4 0 80 104 # 13 1436 450 81 52 6 0 82 110 # 14 1522 434 83 123 27 0 84 273 # 15 1645 415 86 100 40 0 87 240 ``` --- # Mutate ```r model <- denv %>% select(dfs, alt, pH, nit, pho, amm) %>% dplyr::filter(alt > 400) %>% mutate(pH2 = pH + 1) %>% mutate(index = 2*nit + pho + amm) %>% lm(index ~ pH2, data = .) summary(model) # # Call: # lm(formula = index ~ pH2, data = .) # # Residuals: # Min 1Q Median 3Q Max # -117.64 -66.20 -23.96 54.46 212.04 # # Coefficients: # Estimate Std. Error t value Pr(>|t|) # (Intercept) -1243.01 959.57 -1.295 0.218 # pH2 16.84 11.74 1.435 0.175 # # Residual standard error: 93.11 on 13 degrees of freedom # Multiple R-squared: 0.1367, Adjusted R-squared: 0.07033 # F-statistic: 2.059 on 1 and 13 DF, p-value: 0.1749 ``` --- class: inverse, center, middle # Questions? ## <i class="fa fa-question-circle-o" aria-hidden="true"></i> --- # Join <br> -- ```r dfis <- doubs$fish head(denv, 4) # # A tibble: 4 x 11 # dfs alt slo flo pH har pho nit amm oxy bdo # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> # 1 3 934 6.18 84 79 45 1 20 0 122 27 # 2 22 932 3.43 100 80 40 2 20 10 103 19 # 3 102 914 3.64 180 83 52 5 22 5 105 35 # 4 185 854 3.50 253 80 72 10 21 0 110 13 head(dfis, 4) # Cogo Satr Phph Neba Thth Teso Chna Chto Lele Lece Baba Spbi Gogo Eslu Pefl Rham Legi Scer # 1 0 3 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 # 2 0 5 4 3 0 0 0 0 0 0 0 0 0 0 0 0 0 0 # 3 0 5 5 5 0 0 0 0 0 0 0 0 0 1 0 0 0 0 # 4 0 4 5 5 0 0 0 0 0 1 0 0 1 2 2 0 0 0 # Cyca Titi Abbr Icme Acce Ruru Blbj Alal Anan # 1 0 0 0 0 0 0 0 0 0 # 2 0 0 0 0 0 0 0 0 0 # 3 0 0 0 0 0 0 0 0 0 # 4 0 1 0 0 0 0 0 0 0 ``` --- # Join <br> ```r denv <- denv %>% mutate(idSite = 1:nrow(denv)) head(denv, 4) # # A tibble: 4 x 12 # dfs alt slo flo pH har pho nit amm oxy bdo idSite # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> # 1 3 934 6.18 84 79 45 1 20 0 122 27 1 # 2 22 932 3.43 100 80 40 2 20 10 103 19 2 # 3 102 914 3.64 180 83 52 5 22 5 105 35 3 # 4 185 854 3.50 253 80 72 10 21 0 110 13 4 dfis$idSite <- 1:nrow(denv) head(dfis, 4) # Cogo Satr Phph Neba Thth Teso Chna Chto Lele Lece Baba Spbi Gogo Eslu Pefl Rham Legi Scer # 1 0 3 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 # 2 0 5 4 3 0 0 0 0 0 0 0 0 0 0 0 0 0 0 # 3 0 5 5 5 0 0 0 0 0 0 0 0 0 1 0 0 0 0 # 4 0 4 5 5 0 0 0 0 0 1 0 0 1 2 2 0 0 0 # Cyca Titi Abbr Icme Acce Ruru Blbj Alal Anan idSite # 1 0 0 0 0 0 0 0 0 0 1 # 2 0 0 0 0 0 0 0 0 0 2 # 3 0 0 0 0 0 0 0 0 0 3 # 4 0 1 0 0 0 0 0 0 0 4 ``` --- # Join <br> ```r dmerg <- denv %>% inner_join(dfis) head(dmerg) # # A tibble: 6 x 39 # dfs alt slo flo pH har pho nit amm oxy bdo idSite Cogo Satr Phph # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> # 1 3 934 6.18 84 79 45 1 20 0 122 27 1 0 3 0 # 2 22 932 3.43 100 80 40 2 20 10 103 19 2 0 5 4 # 3 102 914 3.64 180 83 52 5 22 5 105 35 3 0 5 5 # 4 185 854 3.50 253 80 72 10 21 0 110 13 4 0 4 5 # 5 215 849 3.18 264 81 84 38 52 20 80 62 5 0 2 3 # 6 324 846 3.50 286 79 60 20 15 0 102 53 6 0 3 4 # # … with 24 more variables: Neba <dbl>, Thth <dbl>, Teso <dbl>, Chna <dbl>, Chto <dbl>, # # Lele <dbl>, Lece <dbl>, Baba <dbl>, Spbi <dbl>, Gogo <dbl>, Eslu <dbl>, Pefl <dbl>, # # Rham <dbl>, Legi <dbl>, Scer <dbl>, Cyca <dbl>, Titi <dbl>, Abbr <dbl>, Icme <dbl>, # # Acce <dbl>, Ruru <dbl>, Blbj <dbl>, Alal <dbl>, Anan <dbl> dim(dmerg) # [1] 30 39 ``` --- # Join <br> ### Several ways of joining data frames [(see documentation)](https://dplyr.tidyverse.org/reference/join.html) ```r dmerg1 <- denv[-1,] %>% inner_join(dfis[-2,]) dim(dmerg1) # [1] 28 39 ``` -- ```r dmerg2 <- denv[-1,] %>% right_join(dfis[-2,]) dmerg3 <- denv[-1,] %>% left_join(dfis[-2,]) dmerg4 <- denv[-1,] %>% full_join(dfis[-2,]) dim(dmerg2) # [1] 29 39 dim(dmerg3) # [1] 29 39 dim(dmerg4) # [1] 30 39 ``` --- # Aggregate <br> -- ```r denv2 <- denv %>% mutate(nit2 = nit >=125) denv2 # # A tibble: 30 x 13 # dfs alt slo flo pH har pho nit amm oxy bdo idSite nit2 # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <lgl> # 1 3 934 6.18 84 79 45 1 20 0 122 27 1 FALSE # 2 22 932 3.43 100 80 40 2 20 10 103 19 2 FALSE # 3 102 914 3.64 180 83 52 5 22 5 105 35 3 FALSE # 4 185 854 3.50 253 80 72 10 21 0 110 13 4 FALSE # 5 215 849 3.18 264 81 84 38 52 20 80 62 5 FALSE # 6 324 846 3.50 286 79 60 20 15 0 102 53 6 FALSE # 7 268 841 4.20 400 81 88 7 15 0 111 22 7 FALSE # 8 491 792 3.26 130 81 94 20 41 12 70 81 8 FALSE # 9 705 752 2.56 480 80 90 30 82 12 72 52 9 FALSE # 10 990 617 4.61 1000 77 82 6 75 1 100 43 10 FALSE # # … with 20 more rows ``` --- # Aggregate <br> ```r denv2$alt2 <- "low" denv2$alt2[denv2$alt>450] <- "medium" denv2$alt2[denv2$alt>750] <- "high" denv2$alt2 <- as.factor(denv2$alt2) denv2 # # A tibble: 30 x 14 # dfs alt slo flo pH har pho nit amm oxy bdo idSite nit2 alt2 # <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <lgl> <fct> # 1 3 934 6.18 84 79 45 1 20 0 122 27 1 FALSE high # 2 22 932 3.43 100 80 40 2 20 10 103 19 2 FALSE high # 3 102 914 3.64 180 83 52 5 22 5 105 35 3 FALSE high # 4 185 854 3.50 253 80 72 10 21 0 110 13 4 FALSE high # 5 215 849 3.18 264 81 84 38 52 20 80 62 5 FALSE high # 6 324 846 3.50 286 79 60 20 15 0 102 53 6 FALSE high # 7 268 841 4.20 400 81 88 7 15 0 111 22 7 FALSE high # 8 491 792 3.26 130 81 94 20 41 12 70 81 8 FALSE high # 9 705 752 2.56 480 80 90 30 82 12 72 52 9 FALSE high # 10 990 617 4.61 1000 77 82 6 75 1 100 43 10 FALSE medium # # … with 20 more rows ``` --- # Aggregate <br> ```r denv2 %>% group_by(nit2) %>% summarize(n()) # # A tibble: 2 x 2 # nit2 `n()` # <lgl> <int> # 1 FALSE 14 # 2 TRUE 16 denv2 %>% group_by(nit2, alt2) %>% summarize(n()) # # A tibble: 5 x 3 # # Groups: nit2 [2] # nit2 alt2 `n()` # <lgl> <fct> <int> # 1 FALSE high 9 # 2 FALSE low 3 # 3 FALSE medium 2 # 4 TRUE low 15 # 5 TRUE medium 1 ``` --- # Aggregate <br> ```r denv2 %>% group_by(nit2) %>% summarize(n()) # # A tibble: 2 x 2 # nit2 `n()` # <lgl> <int> # 1 FALSE 14 # 2 TRUE 16 denv2 %>% group_by(nit2, alt2) %>% summarize(nb_obs = n(), mean_amm = mean(amm), sd_oxy = sd(oxy)) # # A tibble: 5 x 5 # # Groups: nit2 [2] # nit2 alt2 nb_obs mean_amm sd_oxy # <lgl> <fct> <int> <dbl> <dbl> # 1 FALSE high 9 6.56 18.6 # 2 FALSE low 3 0 3.79 # 3 FALSE medium 2 0.5 15.6 # 4 TRUE low 15 37.9 20.6 # 5 TRUE medium 1 0 NA ``` --- # Write files ### Write data frames ```r write.csv(dmerg, "output/dmerg.csv") write_csv(dmerg, "output/dmerg2.csv") ``` -- ### Write any R objects ```r saveRDS(doubs, "output/doubs.rds") # readRDS("output/dmerg.rds") write_rds(doubs, "output/doubs.rds") # read_rds ``` --- class: inverse, center, middle # Data visualization ## <i class="fa fa-bar-chart" aria-hidden="true"></i> --- # Data visualization <br> ## [Material +](https://insileco.github.io/Visualisation-SentinelleNord/#1) ## [bookdown](https://bookdown.org/) --- # Components <br> .pull-left[ - "a chart is worth a thousand words" ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-43-1.png" style="display: block; margin: auto;" /> ] --- # Components <br> .pull-left[ - "a chart is worth a thousand words" - plot window ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-44-1.png" style="display: block; margin: auto;" /> ] --- # Components <br> .pull-left[ - "a chart is worth a thousand words" - plot window - plot region ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-45-1.png" style="display: block; margin: auto;" /> ] --- # Components <br> .pull-left[ - "a chart is worth a thousand words" - plot window - plot region - data ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-46-1.png" style="display: block; margin: auto;" /> ] --- # Components <br> .pull-left[ - "a chart is worth a thousand words" - plot window - plot region - data - axes ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-47-1.png" style="display: block; margin: auto;" /> ] --- # Components <br> .pull-left[ - "a chart is worth a thousand words" - plot window - plot region - data - axes - title ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-48-1.png" style="display: block; margin: auto;" /> ] --- # Components <br> .pull-left[ - "a chart is worth a thousand words" - plot window - plot region - data - axes - titles - legend ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-49-1.png" style="display: block; margin: auto;" /> ] --- # graphics vs grid <br> .center[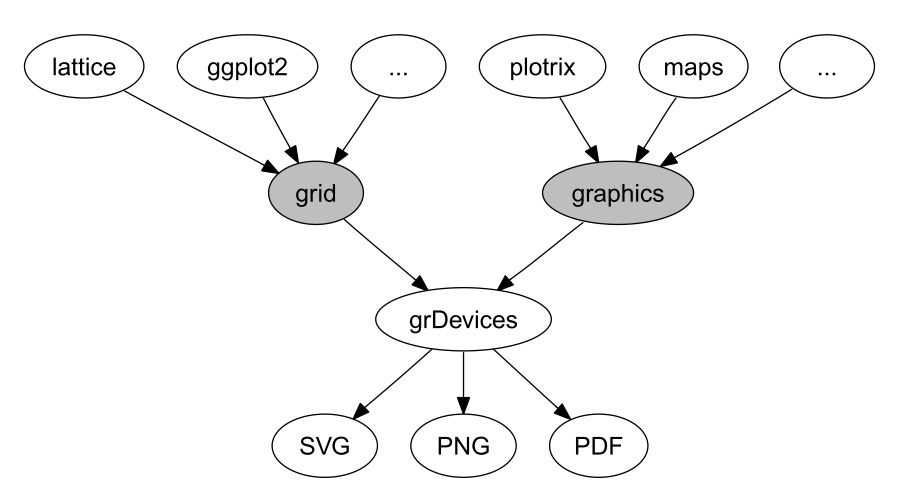] Murrell, P. (2015) [The gridGraphics Package]("https://journal.r-project.org/archive/2015-1/murrell.pdf"). The R Journal. --- # Workflows ### Base plot ```R dev(...) par(...) plot(...) fun1(...) fun2(...) dev.off() ``` <br> ### ggplot2 ```R myplot <- ggplot() + gg_XX1 + gg_XX2 + gg_XX3 myplot ggsave(...) ``` --- # Hundreds of packages <br> ### [List of packages](https://insileco.github.io/wiki/rgraphpkgs/) ### [r-graph-gallery](https://www.r-graph-gallery.com/) ### [data-to-viz](https://www.data-to-viz.com/) ### [Cookbook for R](http://www.cookbook-r.com/Graphs/) --- # Basic plots .pull-left[ ```r # denv as used above plot(x = denv2$alt, y = denv2$amm) ``` <img src="index_files/figure-html/unnamed-chunk-50-1.png" style="display: block; margin: auto;" /> ] .pull-right[ ```r p <- denv2 %>% ggplot(aes(x = alt, y = amm)) + geom_point() p ``` <img src="index_files/figure-html/unnamed-chunk-51-1.png" style="display: block; margin: auto;" /> ] --- # Basic plots .pull-left[ ```r plot(x = denv2$alt, y = denv$amm, type = "l") ``` <img src="index_files/figure-html/unnamed-chunk-52-1.png" style="display: block; margin: auto;" /> ] .pull-right[ ```r p <- denv2 %>% ggplot(aes(x = alt, y = amm)) + geom_line() p ``` <img src="index_files/figure-html/unnamed-chunk-53-1.png" style="display: block; margin: auto;" /> ] --- # Basic plots .pull-left[ ```r par(las = 1) plot(x = denv2$alt, y = denv$amm, type = "l", xlab = "altitude", ylab = "amm") ``` <img src="index_files/figure-html/unnamed-chunk-54-1.png" style="display: block; margin: auto;" /> ] .pull-right[ ```r p <- denv2 %>% ggplot(aes(x = alt, y = amm)) + geom_line() p ``` <img src="index_files/figure-html/unnamed-chunk-55-1.png" style="display: block; margin: auto;" /> ] --- # Basic plots .pull-left[ ```r par(las = 1) plot(x = denv2$alt, y = denv$amm, type = "l", xlab = "altitude", ylab = "amm") ``` <img src="index_files/figure-html/unnamed-chunk-56-1.png" style="display: block; margin: auto;" /> ] .pull-right[ ```r p <- denv2 %>% ggplot(aes(x = alt, y = amm)) + geom_line() + xlab("altitude") + theme_bw() p ``` <img src="index_files/figure-html/unnamed-chunk-57-1.png" style="display: block; margin: auto;" /> ] --- # Basic plots .pull-left[ ```r par(las = 1) plot(x = denv2$alt, y = denv$amm, pch = 19, col = denv2$nit2) ``` <img src="index_files/figure-html/unnamed-chunk-58-1.png" style="display: block; margin: auto;" /> ] .pull-right[ ```r p <- denv2 %>% ggplot(aes(x = alt, y = amm, colour = nit2)) + geom_point() + xlab("altitude") + theme_bw() p ``` <img src="index_files/figure-html/unnamed-chunk-59-1.png" style="display: block; margin: auto;" /> ] --- # Basic plots ```r colors()[1:50] # [1] "white" "aliceblue" "antiquewhite" "antiquewhite1" "antiquewhite2" # [6] "antiquewhite3" "antiquewhite4" "aquamarine" "aquamarine1" "aquamarine2" # [11] "aquamarine3" "aquamarine4" "azure" "azure1" "azure2" # [16] "azure3" "azure4" "beige" "bisque" "bisque1" # [21] "bisque2" "bisque3" "bisque4" "black" "blanchedalmond" # [26] "blue" "blue1" "blue2" "blue3" "blue4" # [31] "blueviolet" "brown" "brown1" "brown2" "brown3" # [36] "brown4" "burlywood" "burlywood1" "burlywood2" "burlywood3" # [41] "burlywood4" "cadetblue" "cadetblue1" "cadetblue2" "cadetblue3" # [46] "cadetblue4" "chartreuse" "chartreuse1" "chartreuse2" "chartreuse3" library(RColorBrewer) ``` --- # Basic plots .pull-left[ ```r par(las = 1) plot(x = denv2$alt, y = denv$amm, pch = 19, xlab = "altitude", ylab = "amm", col = c("salmon1", "turquoise2")[denv2$nit2+1], cex = 2) ``` <img src="index_files/figure-html/unnamed-chunk-61-1.png" style="display: block; margin: auto;" /> ] .pull-right[ ```r p <- denv2 %>% ggplot(aes(x = alt, y = amm, colour = nit2)) + geom_point(size = 3) + xlab("altitude") + theme_bw() p ``` <img src="index_files/figure-html/unnamed-chunk-62-1.png" style="display: block; margin: auto;" /> ] --- # Basic plots .pull-left[ ```r par(las = 1, mar = c(4,4,1,1)) plot(x = denv2$alt, y = denv$amm, pch = 19, xlab = "altitude", ylab = "amm", col = c("salmon1", "turquoise2")[denv2$nit2+1], cex = 1.5) ``` <img src="index_files/figure-html/unnamed-chunk-63-1.png" style="display: block; margin: auto;" /> ] .pull-right[ ```r p <- denv2 %>% ggplot(aes(x = alt, y = amm, colour = nit2)) + geom_point(size = 3) + xlab("altitude") + theme_bw() p ``` <img src="index_files/figure-html/unnamed-chunk-64-1.png" style="display: block; margin: auto;" /> ] --- # Export your figure <br> ```R png("output/figb.png", unit = "in", width = 6, height = 5, res = 300) par(las = 1, mar = c(4,4,1,1)) plot(x = denv2$alt, y = denv$amm, pch = 19, xlab = "altitude", ylab = "amm", col = c("salmon1", "turquoise2")[denv2$nit2+1], cex = 1.5) dev.off() ``` --- # Export your figure <br> ```R p <- denv2 %>% ggplot(aes(x = alt, y = amm, colour = nit2)) + geom_point(size = 3) + xlab("altitude") + theme_bw() ggsave("output/figg.png", unit = "in", width = 6, height = 5) ``` --- # Hints <br> ### Base plots - remove all components and add one layer at a time - use `?par` as well as the help of the functions you'll use <br> ### ggplot2 - https://ggplot2.tidyverse.org/ - find the geom (`gg_XXX`) you need: [ggplot2 extensions](https://www.ggplot2-exts.org/) --- # Other graphs? <br> ## [Cookbook for R](http://www.cookbook-r.com/Graphs/) ## [r-graph-gallery](https://www.r-graph-gallery.com/) --- # Boxplots `amm` vs in `denv2` -- ### Baseplot - `boxplot()` - `par()` -- ### ggplot2 - `geom_boxplot()` --- # Boxplots .pull-left[ ```r par(las = 1) boxplot(oxy~alt2, data = denv2) ``` <img src="index_files/figure-html/unnamed-chunk-65-1.png" style="display: block; margin: auto;" /> ] .pull-right[ ```r p <- denv2 %>% ggplot(aes(x = alt2, y = oxy)) + geom_boxplot() p ``` <img src="index_files/figure-html/unnamed-chunk-66-1.png" style="display: block; margin: auto;" /> ] --- # Custom plot <br> - Let's create a 3-panels figure ```r mat <- matrix(c(1,1,2,3), 2, 2) mat # [,1] [,2] # [1,] 1 2 # [2,] 1 3 ``` --- # Custom plot - 3 panels <br> .pull-left[ ```R layout(mat, widths = c(1.5, 1)) layout.show(3) ``` ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-68-1.png" style="display: block; margin: auto;" /> ] --- # Custom plot - 3 panels <br> .pull-left[ ```R layout(mat, widths = c(1.1, 1)) par(las = 1, mar = c(4,4,1,1)) #-- Panel 1 plot(1,1) #-- Panel 2 plot(x = denv2$alt, y = denv$amm, pch = 19, xlab = "altitude", ylab = "amm", col = c("salmon1", "turquoise2")[denv2$nit2+1], cex = 1.2) #-- Panel 3 boxplot(amm~alt2, data = denv2, ylim = c(0, 80)) ``` ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-69-1.png" style="display: block; margin: auto;" /> ] --- # Custom plot - Panel 1 .pull-left[ ```R plot(denv2$pho, -denv2$alt) ``` ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-70-1.png" style="display: block; margin: auto;" /> ] --- # Custom plot - Panel 1 .pull-left[ ```R par(las = 1, bty = "l") plot(denv2$pho, -denv2$alt, ann = FALSE, axes = FALSE, type = "n") ``` ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-71-1.png" style="display: block; margin: auto;" /> ] --- # Custom plot - Panel 1 .pull-left[ ```R par(las = 1, bty = "l") plot(denv2$pho, -denv2$alt, ann = FALSE, axes = FALSE, type = "n", xlim = c(0, 500), ylim = c(-1000, 0)) points(denv2$pho, -denv2$alt, pch = 19, cex = 1.1) ``` ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-72-1.png" style="display: block; margin: auto;" /> ] --- # Custom plot - Panel 1 .pull-left[ ```R par(las = 1, bty = "l") plot(denv2$pho, -denv2$alt, ann = FALSE, axes = FALSE, type = "n", xlim = c(0, 500), ylim = c(-1000, 0)) lines(denv2$pho, -denv2$alt, lwd = 1.5) axis(1) axis(2, at = seq(-1000, 0, 200), labels = abs(seq(-1000, 0, 200))) ``` ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-73-1.png" style="display: block; margin: auto;" /> ] --- # Custom plot - Panel 1 .pull-left[ ```R par(las = 1, bty = "l") plot(denv2$pho, -denv2$alt, ann = FALSE, axes = FALSE, type = "n", xlim = c(0, 500), ylim = c(-1000, 0)) lines(denv2$pho, -denv2$alt, lwd = 1.5) axis(1) axis(2, at = seq(-1000, 0, 200), labels = abs(seq(-1000, 0, 200))) title(main = "A", xlab = "Phosphorous", ylab = "Altitude") ``` ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-74-1.png" style="display: block; margin: auto;" /> ] --- # Custom plot ```r png("output/figure1.png", unit = "in", width = 7, height = 6, res = 300) layout(mat, widths = c(1.2, 1)) par(mar = c(4,4,1,1)) #-- Panel 1 par(las = 1, bty = "l", xaxs = "i", yaxs = "i") plot(denv2$pho, -denv2$alt, ann = FALSE, axes = FALSE, type = "n", xlim = c(0, 500), ylim = c(-1000, -100)) lines(denv2$pho, -denv2$alt, lwd = 1.5) axis(1) axis(2, at = seq(-1000, 0, 200), labels = abs(seq(-1000, 0, 200))) title(main = "A", xlab = "Phosphorous", ylab = "Altitude") #-- Panel 2 par(bty = "o", xaxs = "r", yaxs = "r") plot(x = denv2$alt, y = denv$amm, pch = 19, xlab = "altitude", ylab = "amm", col = c("salmon1", "turquoise2")[denv2$nit2+1], cex = 1.5) #-- Panel 3 boxplot(oxy~alt2, data = denv2) ``` --- # Custom plot .center[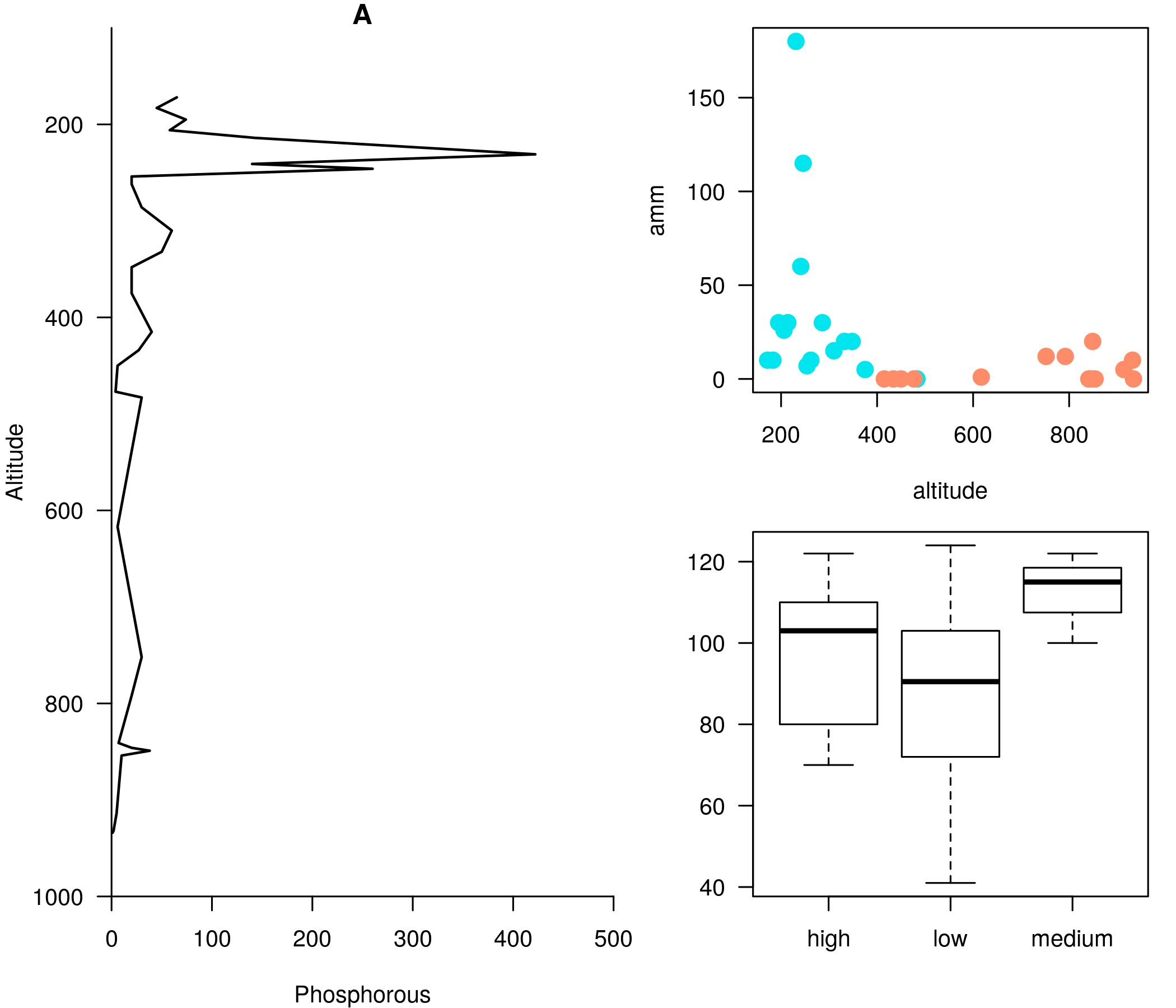] --- # Custom plot <br> - Another example: https://insileco.github.io/Visualisation-SentinelleNord/#33 - See https://insileco.github.io/VisualiseR/ - Partition with ggplot2 - [article on sthda](http://www.sthda.com/english/wiki/ggplot2-facet-split-a-plot-into-a-matrix-of-panels) - [see patchwork](https://github.com/thomasp85/patchwork) --- class: inverse, center, bottom background-image: url(https://upload.wikimedia.org/wikipedia/commons/thumb/a/ad/BlankMap-World_gray.svg/1405px-BlankMap-World_gray.svg.png) background-size: contain # Mapping with R  <br> --- # Overview of the section <br> <img src="images/todo.png" style="float:right;width:170px;margin: 0px 0px"> 1. Why mapping in R? 2. Spatial data & Coordinate reference systems 3. Vector data with **`sf`** 4. Raster data with **`raster`** 5. Thematic maps 6. Interactive maps 7. Questions, discussion, and use of your data --- # Packages for this section <br> ```r library(sf) # spatial vector data library(raster) # spatial raster data library(RColorBrewer) # colors library(mapview) # interactive maps ``` --- # Why mapping in Ecology? .pull-left[ 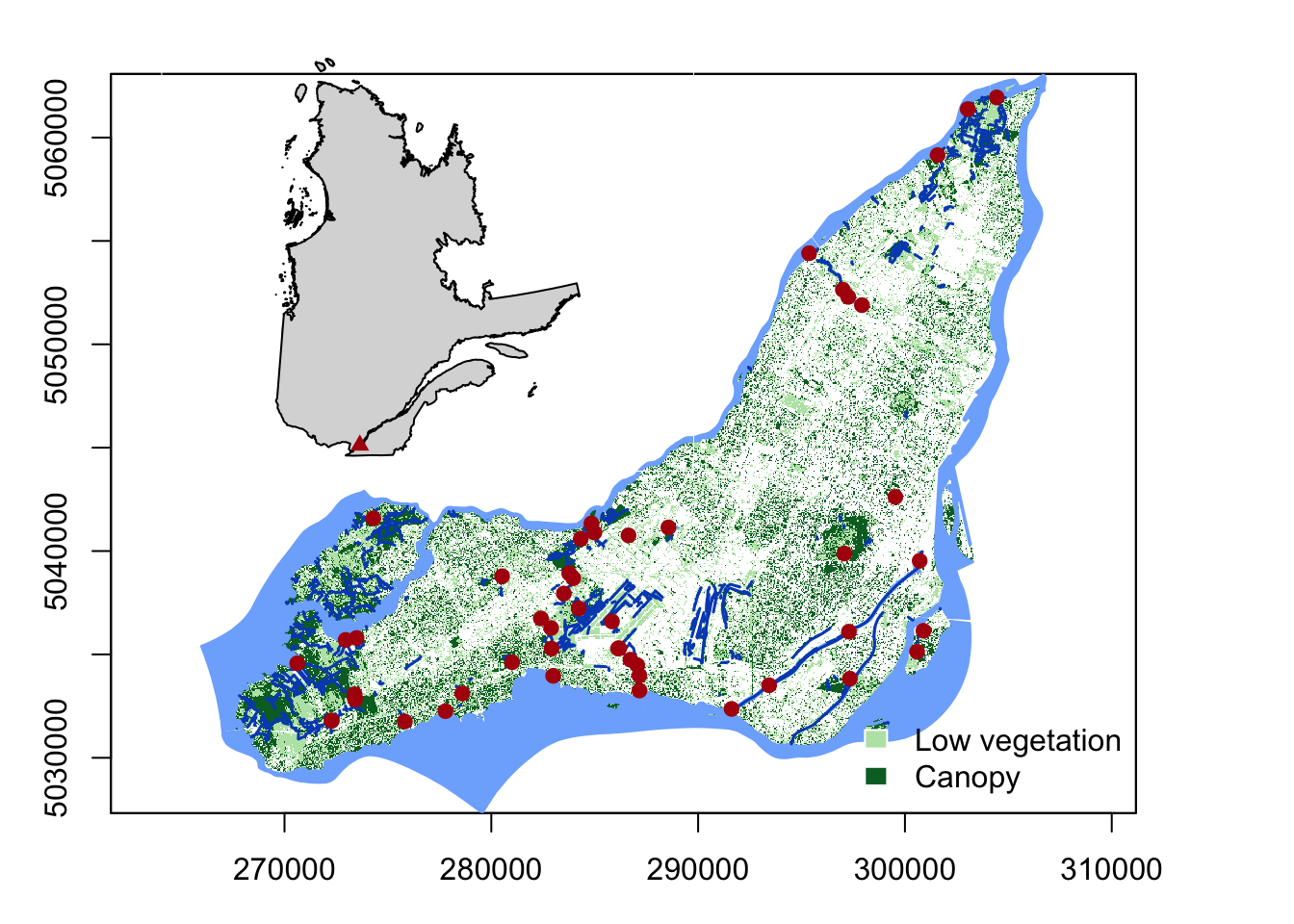 > [R script for this map](https://mhbrice.github.io/Rspatial/Rspatial_script.html) ] .pull-left[ 1. show where your plots are 2. show the spatial distribution of your variables 3. show results of spatial analyses ] --- # Why using R for mapping? 1. Open-source, free 2. Workflow and reproducibility 3. Quite efficient - well-defined spatial classes - can read/write/convert many formats 4. Can also be used for spatial data manipulation and analysis as a GIS [](https://mhbrice.github.io/Rspatial/Rspatial_script.html) --- class: inverse, center, middle # Spatial data --- # Vector data <br><br> .center[ 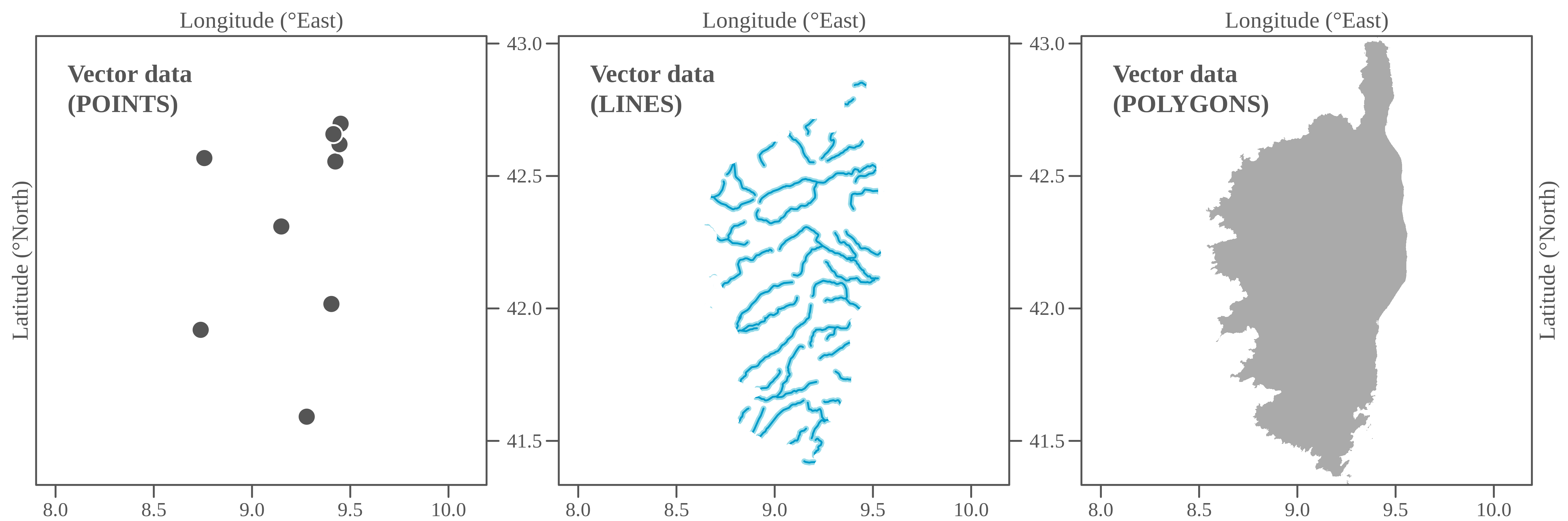 ] --- # Raster data <br><br> .center[  ] --- # Geospatial data in R .column-left[ #### Abiotic data - [`raster`](https://cran.r-project.org/web/packages/raster/index.html) - [`marmap`](https://github.com/ericpante/marmap) - [`rnoaa`](https://github.com/ropensci/rnoaa) - [`rWBclimate`](https://github.com/ropensci/rWBclimate) - [`sdmpredictors`](https://cran.r-project.org/web/packages/sdmpredictors/index.html) ] .column-center[ #### Biotic data - [`rgbif`](https://github.com/ropensci/rgbif) - [`robis`](https://github.com/iobis/robis) - [`spocc`](https://github.com/ropensci/spocc) ] .column-right[ #### Base maps - [`ggmap`](https://github.com/dkahle/ggmap) - [`mregions`](https://github.com/ropenscilabs/mregions) - [`osmdata`](https://github.com/ropensci/osmdata) - [`raster`](https://cran.r-project.org/web/packages/raster/index.html) - [`rnaturalearth`](https://github.com/ropenscilabs/rnaturalearth) ] --- class: inverse, center, middle # Coordinate reference systems .center[ 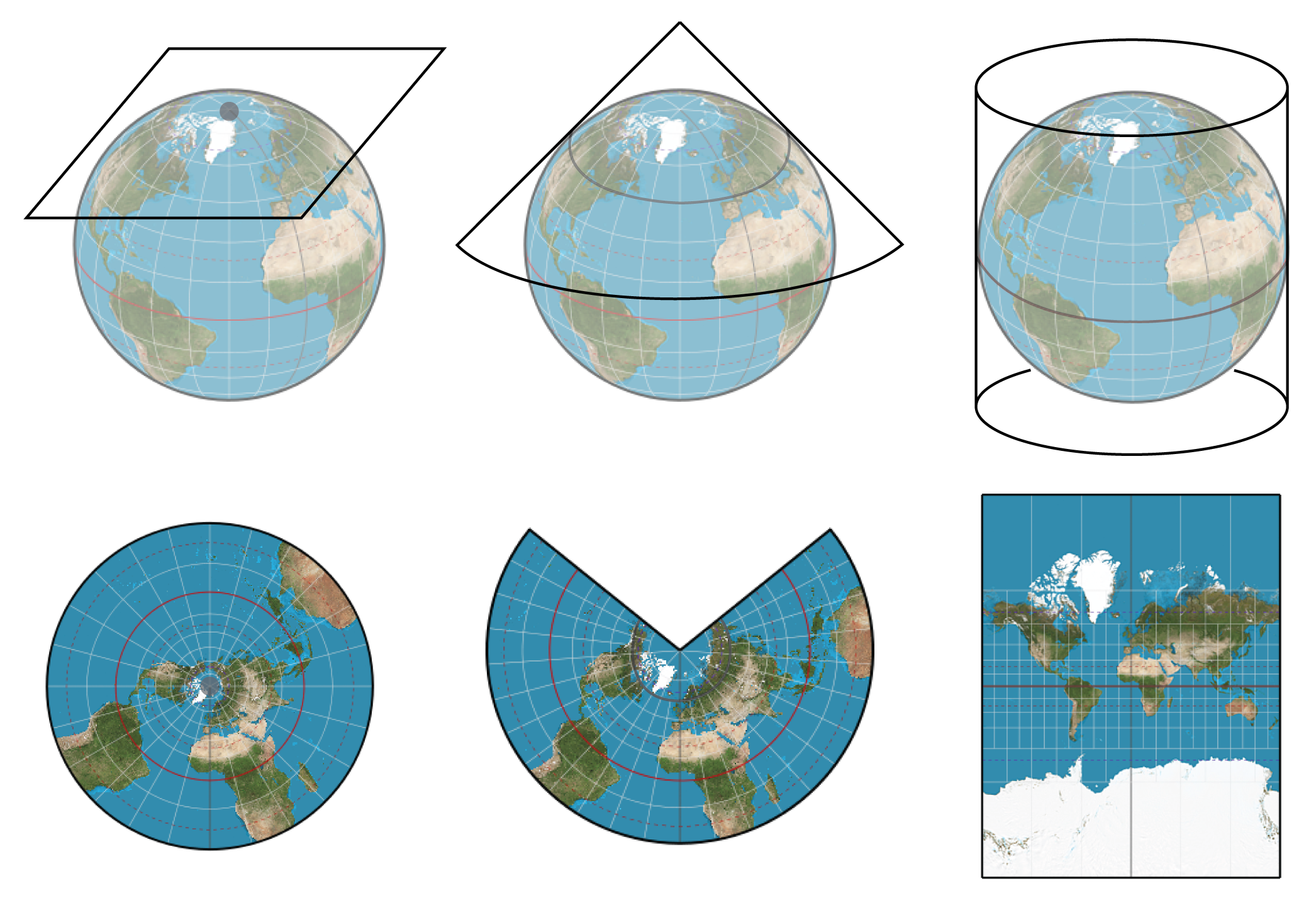 ] --- # Geographic vs projected CRS <img src="index_files/figure-html/canada-1.png" width="648" style="display: block; margin: auto;" /> > [What are geographic coordinate systems?](http://desktop.arcgis.com/en/arcmap/10.3/guide-books/map-projections/about-geographic-coordinate-systems.htm) > <br> > [What are projected coordinate systems?](http://desktop.arcgis.com/en/arcmap/10.3/guide-books/map-projections/about-projected-coordinate-systems.htm) --- # Define CRS with EPSG or proj4string Many, many ways to represent the 3-D shape of the earth and to project it in a 2-D plane - each CRS can be defined either by an `EPSG` or a `proj4string` > The EPSG code is a numeric representation of a CRS, while the proj4string reprensents the full set of parameters spelled out in a string: > > <br> > > EPSG `4326`  proj4 `+proj=longlat +ellps=WGS84 +datum=WGS84 +no_defs` > > <br> > > EPSG `32188`  proj4 `+proj=tmerc +lat_0=0 +lon_0=-73.5 +k=0.9999 +x_0=304800 +y_0=0 +ellps=GRS80 +datum=NAD83 +units=m +no_defs` - All geographic files are created with a specific CRS - but it's not always defined! - To find CRS in any format: [Spatial Reference](http://spatialreference.org/) --- class: inverse, center, middle # Vector data with `sf` --- # Intro to Simple Features <br> <img src="https://user-images.githubusercontent.com/520851/34887433-ce1d130e-f7c6-11e7-83fc-d60ad4fae6bd.gif" style="float:right;width:150px;height:150px;"> - .alert[Simple features refers to a formal standard] (ISO 19125-1:2004) that describes how objects in the real world can be represented in computers - .alert[`sf` objects are easy to plot, manipulate, import and export] - Spatial objects are stored as data frames, with the feature geometries stored in list-columns - `tidyverse` friendly - .alert[GREAT documentation!] See [sf vignettes](https://cran.rstudio.com/web/packages/sf/index.html) --- # Intro to Simple Features .center[  ] --- # Intro to Simple Features  > [sf vignette #1](https://cran.r-project.org/web/packages/sf/vignettes/sf1.html) --- # Geospatial data in R Let's create a thematic map of Quebec (our study area) using data ravailable in R using `getData()` from `raster`: - Canadian provincial boundaries - vector data - Climate data - raster data ```r # Create a new directory dir.create("data") can <- getData("GADM", country = "CAN", level = 1, path = "data") can ``` ``` # class : SpatialPolygonsDataFrame # features : 13 # extent : -141.0069, -52.61889, 41.67693, 83.11042 (xmin, xmax, ymin, ymax) # coord. ref. : +proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0 # variables : 10 # names : GID_0, NAME_0, GID_1, NAME_1, VARNAME_1, NL_NAME_1, TYPE_1, ENGTYPE_1, CC_1, HASC_1 # min values : CAN, Canada, CAN.1_1, Alberta, Acadia|Nouvelle-Écosse, NA, Province, Province, 10, CA.AB # max values : CAN, Canada, CAN.9_1, Yukon, Yukon Territory|Territoire du Yukon|Yukon|Yuk¢n, NA, Territoire, Territory, 62, CA.YT ``` --- # Prep vector data Convert from `sp` (`sf`'s ancester) to `sf` objects to facilitate manipulation ```r can_sf <- st_as_sf(can) can_sf[1:3,] ``` ``` # Simple feature collection with 3 features and 10 fields # geometry type: MULTIPOLYGON # dimension: XY # bbox: xmin: -139.0522 ymin: 48.30861 xmax: -88.96722 ymax: 60.00208 # epsg (SRID): 4326 # proj4string: +proj=longlat +datum=WGS84 +no_defs # GID_0 NAME_0 GID_1 NAME_1 VARNAME_1 NL_NAME_1 # 1 CAN Canada CAN.1_1 Alberta <NA> <NA> # 6 CAN Canada CAN.2_1 British Columbia Colombie britannique|New Caledonia <NA> # 7 CAN Canada CAN.3_1 Manitoba <NA> <NA> # TYPE_1 ENGTYPE_1 CC_1 HASC_1 geometry # 1 Province Province 48 CA.AB MULTIPOLYGON (((-111.9534 4... # 6 Province Province 59 CA.BC MULTIPOLYGON (((-123.5406 4... # 7 Province Province 46 CA.MB MULTIPOLYGON (((-90.385 57.... ``` --- # Prep vector data Retrieve Quebec and surrounding provinces ```r neigh <- c("Québec", "Ontario", "Nova Scotia", "New Brunswick", "Newfoundland and Labrador") qc_neigh <- dplyr::filter(can_sf, NAME_1 %in% neigh) # or using base R # qc_neigh <- can_sf[can_sf$NAME_1 %in% neigh,] qc_neigh # Simple feature collection with 5 features and 10 fields # geometry type: MULTIPOLYGON # dimension: XY # bbox: xmin: -95.15598 ymin: 41.67693 xmax: -52.61889 ymax: 63.69792 # epsg (SRID): 4326 # proj4string: +proj=longlat +datum=WGS84 +no_defs # GID_0 NAME_0 GID_1 NAME_1 # 1 CAN Canada CAN.4_1 New Brunswick # 2 CAN Canada CAN.5_1 Newfoundland and Labrador # 3 CAN Canada CAN.7_1 Nova Scotia # 4 CAN Canada CAN.9_1 Ontario # 5 CAN Canada CAN.11_1 Québec # VARNAME_1 NL_NAME_1 TYPE_1 ENGTYPE_1 CC_1 HASC_1 # 1 Nouveau-Brunswick|Acadia <NA> Province Province 13 CA.NB # 2 Newfoundland|Terre-Neuve|Terre-Neuve-et-Labrador <NA> Province Province 10 CA.NF # 3 Acadia|Nouvelle-Écosse <NA> Province Province 12 CA.NS # 4 Upper Canada <NA> Province Province 35 CA.ON # 5 Lower Canada <NA> Province Province 24 CA.QC # geometry # 1 MULTIPOLYGON (((-66.84995 4... # 2 MULTIPOLYGON (((-53.37361 4... # 3 MULTIPOLYGON (((-66.01603 4... # 4 MULTIPOLYGON (((-74.48135 4... # 5 MULTIPOLYGON (((-74.64564 4... ``` --- # Prep vector data Change projection for a better representation of Quebec. > Here, I chose EPSG 32188 which corresponds to NAD83 - MTM zone 9 ```r qc_neigh_prj <- st_transform(qc_neigh, crs = 32188) qc_neigh_prj # Simple feature collection with 5 features and 10 fields # geometry type: MULTIPOLYGON # dimension: XY # bbox: xmin: -1260743 ymin: 4653809 xmax: 1873536 ymax: 7071370 # epsg (SRID): 32188 # proj4string: +proj=tmerc +lat_0=0 +lon_0=-73.5 +k=0.9999 +x_0=304800 +y_0=0 +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=m +no_defs # GID_0 NAME_0 GID_1 NAME_1 # 1 CAN Canada CAN.4_1 New Brunswick # 2 CAN Canada CAN.5_1 Newfoundland and Labrador # 3 CAN Canada CAN.7_1 Nova Scotia # 4 CAN Canada CAN.9_1 Ontario # 5 CAN Canada CAN.11_1 Québec # VARNAME_1 NL_NAME_1 TYPE_1 ENGTYPE_1 CC_1 HASC_1 # 1 Nouveau-Brunswick|Acadia <NA> Province Province 13 CA.NB # 2 Newfoundland|Terre-Neuve|Terre-Neuve-et-Labrador <NA> Province Province 10 CA.NF # 3 Acadia|Nouvelle-Écosse <NA> Province Province 12 CA.NS # 4 Upper Canada <NA> Province Province 35 CA.ON # 5 Lower Canada <NA> Province Province 24 CA.QC # geometry # 1 MULTIPOLYGON (((833789.8 49... # 2 MULTIPOLYGON (((1840715 537... # 3 MULTIPOLYGON (((910807.4 48... # 4 MULTIPOLYGON (((227546.9 49... # 5 MULTIPOLYGON (((214478.2 49... ``` --- # Prep vector data It would take a while to plot because there is a lot of unnecessary details, so we can simplify the shape of the polygons using `st_simplify()`. ```r qc_neigh_simple <- st_simplify(qc_neigh_prj, dTolerance = 1000) # simplify geometry at a 1km scale) ``` --- # Simple plot - default plot of `sf` object creates multiple maps (one per attribute). - useful for exploring the spatial distribution of different variables. ```r plot(qc_neigh_simple) ``` <img src="index_files/figure-html/base_plot-1.png" width="432" style="display: block; margin: auto;" /> --- # Simple plot ```r # retrieve Québec only qc_simple <- dplyr::filter(qc_neigh_simple, NAME_1=="Québec") plot(st_geometry(qc_neigh_simple), col = "grey65") plot(st_geometry(qc_simple), col = "blue3", add = TRUE) ``` <img src="index_files/figure-html/map_1-1.png" width="432" style="display: block; margin: auto;" /> --- # Create a layer of `MULTIPOINTS` - We sampled vegetation in 100 plots across Québec; - We want to plot them with points proportional to their species richness. - Let's create a data frame with coordinates + random values. ```r # Sample random points from our study area sample_pts <- st_sample(x = qc_simple, size = 100) # Create an attribute of fake species richness (with 5 to 50 species per plot) richness <- sample(x = 5:50, size = length(sample_pts), replace = TRUE) sample_pts <- st_sf(sample_pts, richness = richness) sample_pts # Simple feature collection with 100 features and 1 field # geometry type: POINT # dimension: XY # bbox: xmin: -121687.8 ymin: 5002564 xmax: 1178245 ymax: 6809289 # epsg (SRID): 32188 # proj4string: +proj=tmerc +lat_0=0 +lon_0=-73.5 +k=0.9999 +x_0=304800 +y_0=0 +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=m +no_defs # First 10 features: # richness sample_pts # 1 14 POINT (771.6457 6024937) # 2 35 POINT (131842.4 5418371) # 3 22 POINT (373684.6 5445284) # 4 24 POINT (-90667.16 5625926) # 5 32 POINT (57234.65 6036940) # 6 8 POINT (262141 6543959) # 7 38 POINT (165641.7 6185047) # 8 10 POINT (253332.6 6762438) # 9 16 POINT (521013.7 6304468) # 10 42 POINT (223655.9 6388818) ``` --- # Get bioclimatic rasters - Assuming we are interested in the latitudinal temperature gradient, we add a raster of mean annual temperature (`bio1`) as a background to our map. - Let's use a low resolution so it does not take too long to plot. ```r bioclim <- getData("worldclim", var = "bio", res = 10, path = "data") # There are 19 layers in this raster. plot(bioclim) ``` <img src="index_files/figure-html/temp-1.png" width="576" style="display: block; margin: auto;" /> --- # Prep raster data Look at the annual mean temperature raster data, `bio1`: ```r bioclim$bio1 # class : RasterLayer # dimensions : 900, 2160, 1944000 (nrow, ncol, ncell) # resolution : 0.1666667, 0.1666667 (x, y) # extent : -180, 180, -60, 90 (xmin, xmax, ymin, ymax) # coord. ref. : +proj=longlat +datum=WGS84 +ellps=WGS84 +towgs84=0,0,0 # data source : /home/kevcaz/Github/Tutorials/IntroR/data/wc10/bio1.bil # names : bio1 # values : -269, 314 (min, max) ``` -- We need to divide by 10 (because Worldclim temperature data are in °C * 10) ```r temp <- bioclim$bio1/10 temp # class : RasterLayer # dimensions : 900, 2160, 1944000 (nrow, ncol, ncell) # resolution : 0.1666667, 0.1666667 (x, y) # extent : -180, 180, -60, 90 (xmin, xmax, ymin, ymax) # coord. ref. : +proj=longlat +datum=WGS84 +ellps=WGS84 +towgs84=0,0,0 # data source : in memory # names : bio1 # values : -26.9, 31.4 (min, max) ``` --- # Prep raster data - projection Change projection to match with the polygons using `projectRaster()`. - `projectRaster()` requires the PROJ.4 format of the CRS: ```r st_crs(qc_simple)$proj4string # [1] "+proj=tmerc +lat_0=0 +lon_0=-73.5 +k=0.9999 +x_0=304800 +y_0=0 +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=m +no_defs" ``` ```r temp_prj <- projectRaster(temp, crs = st_crs(qc_simple)$proj4string) ``` --- # Prep raster data - crop and mask 1. `crop()` decreases the extent of a raster using the extent of another spatial object. `crop()` expects a `sp` object, so we need to transform the polygon first. 2. `mask()` keeps the raster values only in the area of interest and set the rest to `NA`. ```r temp_crop <- crop(temp_prj, as(qc_simple, "Spatial")) temp_mask <- mask(temp_crop, qc_simple) ``` ```r par(mfrow = c(1,2)) raster::plot(temp_crop, main = "crop") raster::plot(temp_mask, main = "mask") ``` <img src="index_files/figure-html/unnamed-chunk-81-1.png" width="504" style="display: block; margin: auto;" /> --- class: inverse, center, middle # Thematic map --- # Creating a simple layout ```r my.layout <- layout(matrix(1:2, 2), heights = c(1,.2)) layout.show(my.layout) ``` <img src="index_files/figure-html/layout-1.png" width="396" style="display: block; margin: auto;" /> --- # Define color palette Palette of color for the temperature raster using `brewer.pal()` and `colorRampPalette()` ```r display.brewer.pal(11, "RdYlBu") ``` <img src="index_files/figure-html/color-1.png" width="432" style="display: block; margin: auto;" /> ```r mypal1 <- rev(brewer.pal(11, "RdYlBu")) mypal1 # [1] "#313695" "#4575B4" "#74ADD1" "#ABD9E9" "#E0F3F8" "#FFFFBF" "#FEE090" "#FDAE61" # [9] "#F46D43" "#D73027" "#A50026" mypal2 <- colorRampPalette(mypal1) mypal2(16) # [1] "#313695" "#3E60A9" "#5487BD" "#74ADD1" "#98CAE1" "#BCE1ED" "#E0F3F8" "#F4FBD2" # [9] "#FEF4AF" "#FEE090" "#FDBE70" "#FA9857" "#F46D43" "#E04430" "#C62026" "#A50026" mypal <- mypal2(200) ``` --- # Step by step thematic map Plot neighbor provinces as background ```r layout(matrix(1:2, 2), heights = c(1,.2)) par(las = 1, xaxs='i', yaxs='i', mar = c(2,3,0,0)) plot(st_geometry(qc_neigh_simple), col = '#b5cfbd', border = 'grey50', axes = TRUE, graticule = TRUE) # add graticules ``` <img src="index_files/figure-html/unnamed-chunk-82-1.png" width="396" style="display: block; margin: auto;" /> --- # Step by step thematic map Add the temperature raster ```r raster::image(temp_mask, add = TRUE, col = mypal) ``` <img src="index_files/figure-html/unnamed-chunk-83-1.png" width="396" style="display: block; margin: auto;" /> --- # Step by step thematic map Add the Quebec polygon's boundary on top ```r plot(st_geometry(qc_simple), add = TRUE, border = 'grey15', lwd = 1.4) ``` <img src="index_files/figure-html/unnamed-chunk-84-1.png" width="396" style="display: block; margin: auto;" /> --- # Step by step thematic map Add sample points with size proportional to their species richness ```r plot(st_geometry(sample_pts), add = TRUE, pch = 21, bg = "#63636388", col = "grey15", lwd = 1.4, cex = sample_pts$richness/25) # Size proportional to richness ``` <img src="index_files/figure-html/unnamed-chunk-85-1.png" width="396" style="display: block; margin: auto;" /> --- # Step by step thematic map Create a color legend manually ```r val <- range(values(temp_mask), na.rm = TRUE) val # [1] -9.870578 6.487884 mat <- as.matrix(seq(val[1], val[2], length = 200)) image(x = mat[,1], y = 1, z = mat, col = mypal, axes = FALSE, ann = FALSE) axis(1) mtext(side = 1, line = 1.8, text = 'Mean annual temperature (°C)') ``` <img src="index_files/figure-html/unnamed-chunk-86-1.png" width="432" style="display: block; margin: auto;" /> --- # Step by step thematic map Add the color legend under the map ```r par(mar = c(3,4,1,4)) raster::image(x = mat[,1], y = 1, z = mat, col = mypal, axes = FALSE, ann = FALSE) axis(1, cex.axis = .8) ``` <img src="index_files/figure-html/unnamed-chunk-87-1.png" width="396" style="display: block; margin: auto;" /> --- class: inverse, center, middle # Interactive map --- # Interactive map ```r mapView(qc_simple) ``` <div id="htmlwidget-366dcfab22c070ff5d76" style="width:792px;height:432px;" class="leaflet html-widget"></div> <script type="application/json" data-for="htmlwidget-366dcfab22c070ff5d76">{"x":{"options":{"minZoom":1,"maxZoom":100,"crs":{"crsClass":"L.CRS.EPSG3857","code":null,"proj4def":null,"projectedBounds":null,"options":{}},"preferCanvas":false,"bounceAtZoomLimits":false,"maxBounds":[[[-90,-370]],[[90,370]]]},"calls":[{"method":"addProviderTiles","args":["CartoDB.Positron",1,"CartoDB.Positron",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"addProviderTiles","args":["CartoDB.DarkMatter",2,"CartoDB.DarkMatter",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"addProviderTiles","args":["OpenStreetMap",3,"OpenStreetMap",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"addProviderTiles","args":["Esri.WorldImagery",4,"Esri.WorldImagery",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"addProviderTiles","args":["OpenTopoMap",5,"OpenTopoMap",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"createMapPane","args":["polygon",420]},{"method":"addPolygons","args":[[[[{"lng":[-57.1348063073859,-57.157392547156,-57.1200781882042,-57.1348063073859],"lat":[51.3780524333033,51.4005171628702,51.3882796468767,51.3780524333033]}],[{"lng":[-78.9829025280765,-78.9692512174531,-78.9400024382173,-78.9566650381597,-78.9861144980604,-78.9472198481882,-78.978332518083,-78.9305343582399,-78.9311370782364,-78.9588851881465,-78.939392088207,-78.9566650381503,-78.9310913082321,-78.952774048161,-78.9305572482326,-78.9782104480758,-78.9905548080282,-79.0650024377547,-78.9874343880366,-79.0385131778487,-79.0700073177311,-79.0822143576795,-79.0456237778215,-79.0944518976279,-79.1072235075716,-79.085235597662,-79.143890377409,-79.088317867648,-79.1221923775037,-79.0844726576574,-79.1061401375674,-78.9966659479895,-79.0399780278251,-78.9938888479954,-78.9622192381083,-78.8872222883451,-78.9716644280706,-78.9527511581309,-79.0043945279498,-78.9957046479791,-79.025001527877,-79.0666656477163,-79.0491409277777,-79.0181884778954,-79.0364761378299,-78.9944686879806,-79.06055449773,-79.0223693778757,-79.0525970477581,-79.0166015578952,-78.9871826179988,-79.035018917823,-79.0533370977497,-79.0692748976883,-79.1105575575162,-79.1005248975568,-79.1253433174495,-79.1538925173232,-79.1544189473183,-79.1201782174697,-79.0899963376006,-79.0752105676595,-79.0122222879064,-78.9805297880195,-79.0832061776262,-79.0137939478953,-79.1033325175369,-79.0566635077288,-79.0803222676296,-79.0577773977211,-79.0316696178179,-79.0812454176211,-79.0397643977813,-79.0561141977174,-79.0777740476225,-79.0582885676975,-79.1045761075018,-79.0437698377513,-78.9794616679945,-78.9522094680872,-79.0122222878713,-78.9978027279213,-79.0101242078779,-79.1317901573738,-79.040420527759,-79.1083297674765,-79.0011291479058,-79.0377654977658,-79.0361404377698,-79.1038894674923,-79.0477905277228,-79.0730056776178,-78.9505004880785,-79.031677247779,-79.039428707746,-79.0916747975344,-79.0717620776142,-79.1138915974348,-79.0711135876149,-79.1738586371509,-79.163208007198,-79.132225037345,-79.0550003076746,-79.0561752276663,-79.1149902274122,-79.0588912976458,-79.1799926771044,-79.2161102269247,-79.2077789269707,-79.2538909867286,-79.2644042966686,-79.2172241169169,-79.2438888467724,-79.3077773964171,-79.3005523664572,-79.3294448862871,-79.3349990762505,-79.3916702258913,-79.3550033561206,-79.2811279265593,-79.2977905264621,-79.2788925165682,-79.3793945259523,-79.3749999959786,-79.4133300757245,-79.4183349556863,-79.3605346660681,-79.4271850556238,-79.4038925157799,-79.4155578556957,-79.4850006051909,-79.4538879354178,-79.482177725202,-79.4511108354325,-79.4811096152061,-79.4777755652264,-79.5088882349867,-79.4800033552014,-79.5022201450271,-79.4649963353126,-79.4515991154073,-79.502220145024,-79.4733352652427,-79.4927749550913,-79.4755248952178,-79.5311126647775,-79.4893798751081,-79.5299987747782,-79.4954833950511,-79.5400009146918,-79.5055541949692,-79.5083312949443,-79.5261077848039,-79.5100097649275,-79.5305557247595,-79.511131284914,-79.5855560242829,-79.5422210646555,-79.5027770949649,-79.547790524598,-79.5383300746667,-79.5766677843445,-79.5622253344632,-79.5983352641471,-79.5711059543844,-79.6583251935845,-79.6444091737102,-79.7288513128503,-79.5678100543822,-79.4861144950519,-79.4533309852954,-79.4616088852373,-79.4416656453865,-79.3872070257641,-79.3405532760724,-79.2261276167359,-79.0088882377356,-78.9800796478472,-78.9383315979961,-78.9418945279812,-78.8877792381641,-78.8927001981465,-78.8335494983309,-78.75271605856,-78.7116088886627,-78.5784149189579,-78.5470809890165,-78.3167571993763,-78.1923828095159,-77.9148788497369,-77.8477172897746,-77.7916564898031,-77.6963195798431,-77.6267089798683,-77.5541686998912,-77.4176330599247,-77.3361282299405,-77.3501205399379,-77.3028259299459,-77.2365417499559,-77.1622619599651,-77.1211395299697,-77.0820922899733,-77.1985778799604,-77.0862045299727,-77.0644912699748,-76.9937057499803,-76.9634857199826,-76.9797820999813,-76.8814849899873,-76.82981872999,-76.7958984399913,-76.7322235099938,-76.6905517599951,-76.6411132799962,-76.6583328199957,-76.5776977499978,-76.5511175104416,-76.5438842799985,-76.5072250399991,-76.5500848394598,-76.5344467199987,-76.4744415299996,-76.3255615200012,-76.2883300800014,-76.1383361800019,-76.1251831100019,-76.073333740002,-76.063880920002,-76.015014650002,-75.9577789300019,-75.8988037100019,-75.9905548100019,-75.9438858000019,-76.0161132800019,-76.0005645800019,-76.0299987800019,-76.0333252000019,-76.1005554200019,-76.1138916000018,-76.2272186300015,-76.1495513900017,-76.2216644300015,-76.2200012200015,-76.3372192400008,-76.2761077900012,-76.3183364900009,-76.2666702300012,-76.3179016100009,-76.2344436600013,-76.2161254900014,-76.1511077900016,-76.1488876300016,-76.1244735700017,-76.1555557300016,-76.1811142000015,-76.2361145000013,-76.2005004900015,-76.2183303800014,-76.3199996900008,-76.278335570001,-76.3405532800006,-76.3455810500005,-76.3866043100001,-76.3272247300006,-76.3994750999999,-76.3705520600002,-76.4299926799995,-76.3844451900001,-76.4127807599998,-76.3811111500002,-76.4572219799993,-76.40667725,-76.4461135899996,-76.4432983399996,-76.3822021500004,-76.3738861100005,-76.42669678,-76.4849853499992,-76.4533309899997,-76.5105285599988,-76.4977798499991,-76.5880126999974,-76.6122055099969,-76.5905761699974,-76.4927978499993,-76.4710998499996,-76.5222244299988,-76.6538925199958,-76.643844599996,-76.6671295199954,-76.6210937499966,-76.6254882799964,-76.5460815399981,-76.5578002899977,-76.5288848899982,-76.5183105499983,-76.5510253899976,-76.5557861299974,-76.5294189499976,-76.5416641199968,-76.5683364899962,-76.5749969499958,-76.5512084999962,-76.6293945299935,-76.6691589399921,-76.6760024999917,-76.6995849599907,-76.71403502999,-76.7600097699879,-76.7463378899885,-76.8660278299812,-76.8412780799828,-76.8599853499814,-76.8411254899826,-76.9955139199698,-76.9960937499696,-77.0510864299637,-77.0922241199584,-77.1504058799504,-77.138885499952,-77.1738891599468,-77.1510696399501,-77.1996307399426,-77.1855392499447,-77.2360839799361,-77.3327941899163,-77.3948135399013,-77.417175289895,-77.5266647298613,-77.4472045898863,-77.5155410798646,-77.4971923798705,-77.5410766598554,-77.5324935898583,-77.5527801498513,-77.6049804698311,-77.5766754198421,-77.6297149698206,-77.6759185797995,-77.7683334397516,-77.8114318797257,-77.9633331296166,-78.0562286395323,-78.0684204095197,-78.0083160395767,-78.0518188495351,-78.1016311594844,-78.0983038025302,-78.1084518394765,-78.1440048194365,-78.1439819294369,-78.1762313793987,-78.1590042094191,-78.2407607993138,-78.2290039093292,-78.254028319295,-78.2084732093543,-78.2434539793089,-78.2967452992324,-78.2591857892876,-78.3079223592159,-78.3212280291933,-78.3417968691606,-78.3727798491068,-78.3766632091008,-78.3940048190699,-78.3228759791879,-78.425559999011,-78.4873428288869,-78.4268188490094,-78.4467849689724,-78.4288940390073,-78.3950805690689,-78.3462295491542,-78.4239578190178,-78.4806747389047,-78.4607238789448,-78.4929199188761,-78.5816039986747,-78.5656127887115,-78.5939941386425,-78.5837173486671,-78.530120848794,-78.5267867988008,-78.4962310788681,-78.4862060488886,-78.5156478888248,-78.535583498781,-78.5540084787371,-78.4901199288787,-78.5008697488534,-78.561668398715,-78.5495605487422,-78.4973373388604,-78.5073394788376,-78.4406356789754,-78.4321899389933,-78.4749526989088,-78.427719119002,-78.4035720790477,-78.39054107907,-78.3419036891535,-78.3799972490885,-78.3764953590938,-78.3804931590875,-78.404563899044,-78.4006729090504,-78.430671688995,-78.4613647489335,-78.4922256488699,-78.4939880388653,-78.505706788841,-78.535675048774,-78.501785278849,-78.5305557287843,-78.5139770488188,-78.5394439687622,-78.5644454987029,-78.5756835886709,-78.5484008787359,-78.5365066487615,-78.503478998836,-78.5432128887513,-78.5049667388366,-78.5106964088225,-78.4795608488888,-78.5117874088236,-78.4941024788604,-78.4539794889426,-78.4770736688954,-78.4678955089117,-78.4326095589825,-78.393447879055,-78.387054439067,-78.3500976591298,-78.2884521492276,-78.3324584991587,-78.3500976591287,-78.2817993192355,-78.3154907191807,-78.287223819224,-78.2733459492455,-78.2172241193249,-78.2460937492826,-78.2183303793211,-78.2077789293364,-78.1794433593722,-78.0744018594931,-78.1299972494316,-78.1055526694586,-78.136650089424,-78.1644439693906,-78.1394042994203,-78.1699829093831,-78.1188888494436,-78.1672210693865,-78.1916656493546,-78.1211090094402,-78.1466674794096,-78.1225884396203,-78.1511077894035,-78.1272201494317,-78.1277770994305,-78.1027831994591,-78.0522232095118,-78.0422210695221,-78.0183334395453,-78.0350036595289,-77.9711074795884,-77.9593734695989,-77.9727783195866,-77.9222259496286,-77.9966201795652,-78.0149993895476,-77.9888915995718,-77.9888915995714,-77.9438857996105,-77.9333343496188,-77.8766632096627,-77.8793945296604,-77.8494415296818,-77.9172210696309,-77.8711090096657,-77.8294448896954,-77.838317869689,-77.8144683797048,-77.8416747996864,-77.7966690097163,-77.7727813697307,-77.8088912997081,-77.7811126697252,-77.8466796896821,-77.7855529797222,-77.7472228997448,-77.7727813697297,-77.7015991197696,-77.6512756297948,-77.7343749997515,-77.7305297897538,-77.7743987997283,-77.7638854997348,-77.8199996896991,-77.9261093096218,-77.9177780196287,-77.9516677896005,-77.9199981696261,-77.9400024396095,-77.8672256496663,-77.8411102296845,-77.9100036596331,-77.8861083996514,-77.9100036596326,-77.8833312996531,-77.8900146496477,-77.7694473297291,-77.7594451897347,-77.720642089757,-77.7622222897325,-77.733276369749,-77.7644424397309,-77.7416686997442,-77.7521972697378,-77.7905883797145,-77.7822799697194,-77.8066635097039,-77.7911148097136,-77.8266296396899,-77.7638854997299,-77.8088989297012,-77.7455520597405,-77.7433319097416,-77.6949996897682,-77.6998290997653,-77.6555557297879,-77.6655883797832,-77.6172256498064,-77.6805572497761,-77.7255554197517,-77.7261123697517,-77.6905517597712,-77.6750030497789,-77.6649780297841,-77.6266479498023,-77.6555786097888,-77.6249999998034,-77.5927123998175,-77.616043089807,-77.5966796898154,-77.5849990798208,-77.5305786098426,-77.4761199998624,-77.4866561898582,-77.5338745098411,-77.5449829098369,-77.5521850598339,-77.5705566398265,-77.56167602983,-77.5322189298416,-77.511665339849,-77.5704956098263,-77.5700073198259,-77.5244445798438,-77.5122222898485,-77.4711074798627,-77.4866638198572,-77.4494018598694,-77.4327773998748,-77.4233627298775,-77.326599119904,-77.2827758799141,-77.3594436598954,-77.3011093099098,-77.3499984698977,-77.4059753398817,-77.4561080898663,-77.47396087986,-77.3971328698841,-77.4292983998745,-77.4115142798796,-77.3313064599015,-77.3460159298976,-77.290733339911,-77.3297958399016,-77.1618804899366,-77.3369827298997,-77.2687530499157,-77.244270319921,-77.1880187999318,-77.2104186999275,-77.3562469498946,-77.4062499998803,-77.4645843498622,-77.4979171798505,-77.5145797698445,-77.6390609697916,-77.5963516198111,-77.6057281498064,-77.4692687998597,-77.5703124998209,-77.5645828198231,-77.5333328198358,-77.4776000998561,-77.5119781498437,-77.5286483798372,-77.5088500998446,-77.5723953198192,-77.5317687998356,-77.5609359698235,-77.5708312998196,-77.6078109698033,-77.5567703198251,-77.5916671798103,-77.6041641198045,-77.651565549782,-77.652603149781,-77.6890640297624,-77.7265624997413,-77.71093749975,-77.7723998997133,-77.7703170797144,-77.7401046797327,-77.7416686997316,-77.7005157497551,-77.7119827297484,-77.6630172697745,-77.6724014297695,-77.5859374998107,-77.5786514298134,-77.39427184988,-77.5166702298384,-77.5895843498083,-77.6708297697695,-77.7296829197364,-77.7974014296938,-77.8411483796626,-77.8348998996668,-77.7005157497515,-77.6786498997629,-77.6192703197926,-77.6432342497808,-77.6270828197886,-77.7265624997358,-77.6583328197729,-77.7244796797368,-77.5130157498372,-77.7791671797028,-77.8520812996524,-77.8869781496253,-77.8520812996518,-77.974998469549,-78.0895843494294,-78.0833358794361,-78.2244796792543,-78.1932296792963,-77.8703155496334,-77.8442687996535,-77.8244781496662,-77.7807311996959,-77.7505187997154,-77.7598953197092,-77.7208328197333,-77.7088546797402,-77.7151031497362,-77.7598953197087,-77.7421874997193,-77.7703170797008,-77.724998469729,-77.7416686997187,-77.7765655496964,-77.7609329197059,-77.8140640296687,-77.7734374996966,-77.732810969723,-77.6713485697585,-77.6874999997499,-77.6130218497884,-77.705734249739,-77.701560969741,-77.6208343497839,-77.5588531498122,-77.703651429739,-77.7036514297387,-77.6479186997694,-77.6125030497869,-77.5416641198189,-77.4963531498374,-77.4984359698363,-77.6473998997692,-77.5713500998058,-77.5869827297983,-77.4124984698657,-77.5041656498331,-77.5229186998259,-77.5703124998056,-77.6020812997915,-77.7229156497267,-77.7296829197222,-77.7765655496919,-77.7859344496849,-77.9583358795462,-78.0182342494858,-78.0130157494904,-78.0890655494033,-78.0817718494104,-78.1494827293233,-78.1338500993431,-78.1828155492734,-78.14791869932,-78.1494827293157,-78.0942687993864,-78.1369781493319,-78.1067657493705,-78.1119842493634,-78.0505218494368,-78.0666656494179,-78.0953140293836,-78.0911483793884,-78.0005187994904,-78.0474014294403,-78.0374984694518,-77.9604186995309,-77.9083328195791,-77.9286498995601,-77.8833312996001,-77.8973998995878,-77.8333358796399,-77.784896849675,-77.7151031497206,-77.6729202297449,-77.610420229778,-77.575515749795,-77.507812499824,-77.4250030498551,-77.4020843498631,-77.3666686998741,-77.3020858798925,-77.1609344499247,-77.1541671799262,-77.1390609699289,-77.0937499999368,-77.062499999942,-77.0562515299428,-77.0395812999454,-77.0333328199462,-76.9229202299603,-76.9557342499566,-76.888015749964,-76.889579769964,-76.8661498999662,-76.8520812999676,-76.8000030499721,-76.7588500999754,-76.7624969499751,-76.7109374999787,-76.6958312999797,-76.6729202299811,-76.6338500999833,-76.6619796799818,-76.6604156499819,-76.6249999999838,-76.581253049986,-76.5520858799872,-76.5312499999881,-76.5124969499888,-76.4270858799917,-76.3937530499926,-76.339584349994,-76.3083343499946,-76.1854171799968,-76.1125030499977,-76.0901031499979,-76.1145858799977,-76.0270843499985,-76.0187530499985,-75.9849014299988,-75.939582819999,-75.8041686999996,-75.66041565,-75.5651016200001,-75.7437515299998,-75.8265609699996,-75.8723983799994,-75.8625030499994,-75.8041686999997,-75.7713546799997,-75.7828140299997,-75.64427185,-75.62864685,-75.5255203200001,-75.4562530500001,-75.4265670800001,-75.3916702300001,-75.4098968500001,-75.3437500000001,-75.1088485700001,-75.1291656500001,-75.0838546800001,-75.0770797700001,-74.9250030500001,-74.8442688000001,-74.8536453200001,-74.81666565,-74.7421875,-74.76823425,-74.71875,-74.63541412,-74.52916718,-74.64531708,-74.74531555,-74.7098999,-74.39375305,-74.32499695,-74.29426575,-74.26615143,-74.21458435,-74.17708588,-74.16249847,-74.07499695,-73.98593903,-73.95573425,-73.84791565,-73.67500305,-73.60051727,-73.6161499,-73.57551575,-73.54948425,-73.46458435,-73.4458313,-73.3833313,-73.30833435,-73.19010162,-73.20365143,-73.05052185,-73.04948425,-73.01457977,-72.97447968,-72.87916565,-72.8833313,-72.92656708,-72.9057312,-72.86250305,-72.83125305,-72.7895813,-72.79948425,-72.75208282,-72.60676575,-72.61093903,-72.65989685,-72.57968903,-72.6244812,-72.5942688,-72.5859375,-72.66197968,-72.65885162,-72.73906708,-72.71302032,-72.68906403,-72.64375305,-72.63489532,-72.5942688,-72.62291718,-72.58750153,-72.5229187,-72.47135162,-72.4458313,-72.27916718,-72.1859359699999,-72.18489838,-72.1109390299999,-72.2536468499999,-72.1713485699999,-72.2203140299999,-72.2020797699999,-72.1583328199999,-72.1229171799999,-72.0755157499999,-72.0875015299999,-72.0463485699999,-71.9791641199999,-72.0645828199999,-72.1208343499999,-72.1729202299999,-72.24375153,-72.2723998999999,-72.1833343499999,-72.1166686999999,-71.9651031499999,-71.9109344499999,-71.9416656499999,-71.8900985699999,-71.9000015299999,-71.8124999999998,-71.8041686999998,-71.7645797699998,-71.7666702299998,-71.6374969499998,-71.5526046799998,-71.5312499999998,-71.5150985699998,-71.5380172699998,-71.6057281499998,-71.5775985699998,-71.5937499999998,-71.6458358799998,-71.8265609699998,-71.7401046799998,-71.7723998999998,-71.7484359699998,-71.7932281499998,-71.7088546799998,-71.7624969499998,-71.8619842499998,-71.6895828199998,-71.5671844499998,-71.6307296799998,-71.6067657499998,-71.6453170799998,-71.6395797699998,-71.8375015299998,-71.7692718499998,-71.8208312999998,-71.6916656499998,-71.6744842499998,-71.6421890299998,-71.7411498999998,-71.6817703199998,-71.7453155499998,-71.6312484699998,-71.5734329199998,-71.6244811999998,-71.6166686999998,-71.5994796799998,-71.5421829199998,-71.5421829199998,-71.5744781499998,-71.4921874999998,-71.5203170799998,-71.4458312999998,-71.4453124999998,-71.4291686999998,-71.4062499999998,-71.3796844499998,-71.3979186999998,-71.3755187999998,-71.3937530499998,-71.3729171799998,-71.3661498999998,-71.3249969499999,-71.2791671799999,-71.21510315,-71.22447968,-71.1921844500001,-71.1750030500001,-71.1807327300001,-71.1479187000002,-71.0895843500004,-71.0437469500006,-70.985420230001,-70.9109344500015,-70.9286499000013,-70.8354187000022,-70.769271850003,-70.7354202300035,-70.768753050003,-70.7458343500034,-70.7395858800035,-70.7187500000038,-70.7140655500039,-70.6916656500043,-70.6442718500052,-70.6708297700046,-70.6187515300057,-70.5895843500064,-70.5374984700078,-70.5557327300073,-70.5130157500086,-70.4895858800094,-70.4598999000104,-70.4062500000125,-70.4265670800118,-70.3645858800145,-70.4062500000127,-70.3734359700141,-70.3375015300159,-70.3432312000156,-70.3046875000177,-70.300003050018,-70.2317657500224,-70.2291641200225,-70.1312484700304,-70.1411514300294,-70.1104202300322,-70.102600100033,-70.1536483800282,-70.0838546800345,-70.0734329200354,-70.1312484700298,-70.1411514300289,-70.0791702300348,-70.0578155500368,-69.9916687000445,-69.9020843500568,-69.8296890300693,-69.8744812000615,-69.8958358800579,-69.9265670800533,-69.8442688000667,-69.8328170800689,-69.7583313000839,-69.7296829200902,-69.7395858800878,-69.6359329201139,-69.6317672701154,-69.6833343501017,-69.6703109701056,-69.6296844501168,-69.6453170801125,-69.6083297701234,-69.510932920156,-69.4567718501767,-69.4828109701662,-69.4494781501791,-69.4171829201929,-69.4369812001841,-69.3713531502138,-69.3755188002115,-69.402084350199,-69.3630218502166,-69.3708343502127,-69.4000015301992,-69.4224014301891,-69.4109344501939,-69.5307312001461,-69.5744781501313,-69.5505218501391,-69.5567703201369,-69.625000000116,-69.6192703201174,-69.6973953200965,-69.6692657501034,-69.6869812000987,-69.6479187001088,-69.6682281501032,-69.6276016201142,-69.6255188001147,-69.6619796801047,-69.6541671801066,-69.6973953200956,-69.675521850101,-69.7791671800774,-69.8036499000725,-69.774482730078,-69.7130203200912,-69.757812500081,-69.7109375000914,-69.7140655500905,-69.6630172701028,-69.6911468500958,-69.6875000000966,-69.7223968500883,-69.7619781500795,-69.6630172701023,-69.6765670800987,-69.6380157501087,-69.6265640301117,-69.5838546801241,-69.5984344501195,-69.6479187001056,-69.5875015301226,-69.6328125001093,-69.6026001001176,-69.6541671801032,-69.6109390301148,-69.7104187000892,-69.7166671800877,-69.8223953200665,-69.8328170800644,-69.775520320075,-69.7819137600736,-70.3208313000147,-70.2755203200171,-70.5890655500053,-70.6187515300047,-70.7958297700019,-70.9958343500004,-71.0369796800002,-70.9812469500005,-70.7770843500021,-70.6333313000043,-70.6464843700041,-70.5683364900057,-70.3682174700124,-70.3109130900151,-70.3021774300156,-70.2169113200207,-70.1677246100243,-70.1772155800235,-70.076416020032,-70.0065155000394,-69.8911056500546,-69.7205810500863,-69.7083663900887,-69.7694473300756,-69.7603225700774,-69.6666412400985,-69.6666030900984,-69.5460815401328,-69.5944213901176,-69.5708770801244,-69.605552670114,-69.5521850601299,-69.5827789301204,-69.5566635101282,-69.5744476301226,-69.5839157101199,-69.6672210700968,-69.592269900117,-69.595275880116,-69.5066680901442,-69.5016632101456,-69.5588913001261,-69.5947113001154,-69.5932617201159,-69.6399993901029,-69.7388916000793,-69.7482833900771,-69.6844482400911,-69.7055587800861,-69.6560974100978,-69.6339111301032,-69.6666870100947,-69.6211090101063,-69.6261215201047,-69.6311111501033,-69.7266693100807,-69.7746734600708,-69.6961059600871,-69.6641235400947,-69.6204834001057,-69.584396360116,-69.5350036601312,-69.4315795901688,-69.438293460166,-69.3511123702036,-69.3571624802002,-69.326568600215,-69.3122253402221,-69.2916870102332,-69.2455444302588,-69.2367782602634,-69.2716064502436,-69.2694091802444,-69.2272186302678,-69.2451782202574,-69.2469635002561,-69.2910766602319,-69.2894439702323,-69.3577804601991,-69.3750076301905,-69.4152832001734,-69.4099884001759,-69.4405517601637,-69.4932861301439,-69.5250015301328,-69.4983367901411,-69.4566040001564,-69.4399871801624,-69.3750000001897,-69.3438873302035,-69.4044418301762,-69.4361190801629,-69.4899902301436,-69.4381103501607,-69.4758224501464,-69.5183105501322,-69.5444946301229,-69.6211090101011,-69.6470947300947,-69.6511077900935,-69.6811142000867,-69.6959228500839,-69.6815795900872,-69.6644287100911,-69.6578369100932,-69.6887893700856,-69.6983184800842,-69.7222213700792,-69.7247924800783,-69.7700195300698,-69.7679977400697,-69.8438262900571,-69.8662109400536,-69.8511123700555,-69.8672714200531,-69.894241330049,-69.8088607800623,-69.7866821300654,-69.8266677900586,-69.8899993900492,-69.8788909900505,-69.9740371700382,-70.0257034300327,-70.0655517600287,-70.0149993900334,-69.9067916900464,-69.9167175300446,-69.8422241200553,-69.832984920057,-69.8166656500594,-69.7816772500655,-69.7875976600643,-69.8361206100561,-69.8658447300514,-69.8122253400594,-69.6777801500867,-69.6419372600947,-69.6118698101033,-69.533889770126,-69.5327758801262,-69.5134887701325,-69.5005569501366,-69.4738922101462,-69.4666748001486,-69.4428100601579,-69.4072189301719,-69.4143066401694,-69.3865966801812,-69.2954101602241,-69.253479000247,-69.227294920262,-69.2294464102604,-69.2021484402772,-69.2077789302732,-69.1433105503157,-69.0565719603807,-69.0449829103905,-69.0386962903955,-68.9877777104408,-69.0105590804192,-68.9771118204501,-68.8611145005717,-68.7615661606995,-68.7416687007265,-68.6322021508998,-68.631103520901,-68.5816650409903,-68.5366668710759,-68.5216064511073,-68.502777101144,-68.4711990412142,-68.4716796912119,-68.3944473313967,-68.4022216813751,-68.3499755915112,-68.38277435142,-68.3361954162651,-68.3194427515901,-68.3110961916133,-68.3354803628885,-68.3255538915713,-68.3411254915216,-68.2708129917235,-68.2938919116506,-68.2366638218277,-68.2073593119263,-68.1866684019968,-68.2130432118913,-68.3084487915853,-68.3133316015667,-68.341079711489,-68.3050003115853,-68.2986221316095,-68.2427597017829,-68.249656681765,-68.1864013719799,-68.1657943720598,-68.1842041019989,-68.1483306921314,-68.1248168922209,-68.0681686424444,-68.0494613625251,-68.024444582633,-68.0074539227188,-67.9566650429557,-67.9777755728523,-67.9516677929785,-67.9705581728836,-67.93666840305,-67.9243774431146,-67.9022216832278,-67.9171752931417,-67.9527740529587,-67.9282836930821,-67.9329681430531,-67.9072265631841,-67.9238891630962,-67.8905029332662,-67.8721923833674,-67.8577880934461,-67.8477783234988,-67.8750000033471,-67.860000613427,-67.8983306932104,-67.9672241228619,-68.1008834822697,-68.1012191822626,-68.0923919722991,-68.0006103526918,-67.941101072988,-67.8633346633996,-67.8522186334605,-67.8544464134527,-67.8061447137418,-67.831665043592,-67.8422241235437,-67.8283081136313,-67.7933120738441,-67.8022232137954,-67.7494201741312,-67.7261123742854,-67.7432861341671,-67.7166671843504,-67.7183303843354,-67.6832885745815,-67.7155761743502,-67.7121582043705,-67.7277832042653,-67.7288818442493,-67.7055587844075,-67.7516632140883,-67.7116088943539,-67.7316665642134,-67.6894455045093,-67.6427993848502,-67.7100219743367,-67.7011108443915,-67.6766662645645,-67.5838851953095,-67.5438919156506,-67.4561157264832,-67.3955535971038,-67.421463016826,-67.3765869173032,-67.3266677978855,-67.268333448604,-67.2410888789495,-67.2183685392562,-67.1767425598473,-67.1788864198278,-67.0928726311009,-67.0483932618463,-67.0794067513497,-67.0543714194533,-67.0577469017119,-67.0377807720467,-67.05109101953,-67.0133361924662,-66.963317883414,-66.9300003240531,-66.9402694838619,-66.8705520752483,-66.9000244246265,-66.852218645606,-66.8533325356029,-66.8316040160807,-66.8116684165169,-66.7933273469489,-66.8072204766481,-66.7633361976727,-66.7322235284449,-66.6855545196204,-66.6444473507364,-66.6194458214244,-66.6605530003371,-66.6395263909237,-66.6138916216411,-66.613288901719,-66.5905533023786,-66.5644455231772,-66.5776977728088,-66.5538940635287,-66.5272217044006,-66.5474090837748,-66.5161133047645,-66.4647293364958,-66.4699402163035,-66.4439087172077,-66.4349975875522,-66.4556885068211,-66.4755554461578,-66.4466629271724,-66.4594040167684,-66.4505920670865,-66.4249878179877,-66.4211120881447,-66.3843994394954,-66.3786010997435,-66.3338013014816,-66.3483353008723,-66.2944641430706,-66.2294464458641,-66.1601486590167,-66.1561127092374,-66.1153030812472,-66.0632935038553,-66.0133362266079,-66.0272217258551,-66.0004883273877,-65.9755554688196,-65.9972076875248,-65.9772186786806,-66.0000000473538,-65.9266663117358,-65.9555588298179,-66.0021820570533,-66.0132599363751,-65.9500122600749,-65.9550171398053,-65.9100037125914,-65.9166641721935,-65.8872070840576,-65.8911133339458,-65.8215942985751,-65.8816681446218,-65.9377747110918,-65.9382935111372,-65.9777756187383,-66.0493774846854,-66.0405273852022,-66.0205536363152,-66.0585937942437,-66.018447926456,-66.0322647557347,-66.060554544203,-66.0636139339996,-66.0933227924351,-66.1022186720161,-66.0788879832418,-66.0700073637765,-66.0322266058555,-66.0355530257075,-65.9956055179948,-65.958847100114,-65.9326172417604,-65.8789825951727,-65.8655319760929,-65.82491307877,-65.8212280891632,-65.8005982006341,-65.7881546614851,-65.7760315524224,-65.9101486732565,-65.9215546225042,-66.0022201976875,-65.8311081486708,-65.8671875563181,-65.8372192983109,-65.7703247731119,-65.6622238915189,-65.6628266115612,-65.7583313640383,-65.7850037220992,-65.6703797110705,-65.7727051430413,-65.7716065131484,-65.7238922767855,-65.6860962598479,-65.6272202247638,-65.6055527465795,-65.5722199295654,-65.5643921703145,-65.5022202361676,-65.4004441264864,-65.3619767207258,-65.497779936651,-65.5171967447315,-65.5377808428287,-65.5833359585972,-65.5471802619705,-65.5811157988326,-65.5919800579014,-65.5387726628138,-65.5385132628733,-65.4913407274767,-65.4466629919152,-65.4572144508821,-65.3533936520984,-65.3566666617823,-65.3083344573768,-65.3388901838568,-65.4066697162436,-65.4994431366993,-65.5444413024371,-65.5238495742987,-65.633331374293,-65.6549683325091,-65.6343994842755,-65.7516632749475,-65.7310791665487,-65.7155533477351,-65.7127991380113,-65.6399994640474,-65.7379644183434,-65.7371826861125,-65.7583008445132,-65.7388916660094,-65.7380812813511,-65.7190857575716,-65.7549973148091,-65.71055609828,-65.7482910853919,-65.6955567095356,-65.7216644975008,-65.6733322813955,-65.6694412916714,-65.6572190027255,-65.6294174950846,-65.597778397692,-65.5577774812678,-65.6022187074441,-65.6055451172033,-65.5822144393047,-65.5033341364832,-65.4705582597541,-65.5572205416002,-65.5505524523057,-65.48666390844,-65.4772187194457,-65.5194474152541,-65.5072251265087,-65.5566635917713,-65.5716019504969,-65.5333710541006,-65.6392517845927,-65.5928192887072,-65.5876313091276,-65.5509034025477,-65.5032959870706,-65.5350037440499,-65.4954834878956,-65.3105546078036,-65.3349991850205,-65.2883149306419,-65.3522187330395,-65.3933335385729,-65.4516678823213,-65.4379883738646,-65.4138871164366,-65.4271851550979,-65.3383179749979,-65.3877793394352,-65.4961090980066,-65.510109036674,-65.4433747234602,-65.4719239205555,-65.5155564261704,-65.5141983863584,-65.441101163819,-65.4052124976243,-65.3844605499394,-65.419983006076,-65.3984452284416,-65.2561112650996,-65.3922959391702,-65.2988892700377,-65.2880860514187,-65.26540386419,-65.16333020772,-65.17833722555,-65.1066666955684,-64.9522096299743,-65.0977174269427,-65.1544419588741,-65.1472169299918,-65.1727753964611,-65.2938843907548,-65.3894577995143,-65.4377747544159,-65.4321900350913,-65.5149460667903,-65.4911423591507,-65.4821778200973,-65.5327759651273,-65.537780844766,-65.5032959881609,-65.4902344595494,-65.4588852827516,-65.4849854401537,-65.4266663563127,-65.4305573459438,-65.4043885288341,-65.3688508127705,-65.3827744512213,-65.3527833047001,-65.3944855699643,-65.3694459028061,-65.3538819446772,-65.343330485886,-65.3222046985212,-65.2595826361811,-65.2611085160372,-65.212219362416,-65.1613770792368,-65.1321641334946,-65.1033326576814,-65.0972291386377,-65.159446849602,-65.1694413482683,-65.1816636365365,-65.2272187504543,-65.2338431833529,-65.2344437795209,-65.2794419437135,-65.3094483500515,-65.2961426917292,-65.2465992646585,-65.2748795744526,-65.2005540141357,-65.2373048093121,-65.1957703849104,-65.159629949833,-65.1536179907575,-65.1269456345621,-65.1649781592643,-65.1838914266429,-65.1705552384716,-65.1939088152086,-65.2324677700786,-65.2143479625482,-65.1566773804582,-65.1761170677748,-65.142219672649,-65.1402131430361,-65.0006868043724,-65.12333692553,-65.1198121461336,-65.0752770824669,-65.0741654830482,-65.082779071676,-65.1348954539584,-65.1333314343802,-65.1187516665806,-65.0963517598918,-65.0979157897055,-65.0645829647552,-65.0744782932853,-65.0479203774373,-65.0213548317928,-65.0708314439715,-65.033332970083,-64.9567720127419,-65.0057298347435,-64.9770814595868,-64.965103311574,-64.9661485415903,-64.9208299396881,-64.8900987452782,-64.8974000740309,-64.8526002824156,-64.8874971259005,-64.9473954850036,-64.9562532136434,-64.9187471204226,-64.8921892052053,-64.8494798635428,-64.8026048728613,-64.8349000864786,-64.8432313850841,-64.8125001910674,-64.8020860732358,-64.7428348499057,-64.7349016370852,-64.7145845615371,-64.604164356785,-64.6380159788891,-64.6895830370479,-64.7286455285958,-64.6229174125432,-64.5229265872223,-64.7074129328758,-64.8622590908112,-64.8639451903276,-64.7498781329939,-64.7455598937906,-64.6824877076535,-64.6024706362394,-64.5937197180135,-64.6330721285159,-64.7672960388673,-64.7079851313749,-64.7427141337107,-64.9236451884387,-64.8402330334683,-64.7471010322862,-64.7474214620984,-64.6931230738481,-64.6622316707439,-64.6694185489648,-64.6267931387949,-64.6837770753957,-64.7561647498504,-64.7665559875976,-64.7369463137035,-64.7780839951327,-64.8252260155715,-64.7884752327327,-64.8139879176008,-64.7899858521764,-64.8102037379712,-64.749435620227,-64.8249208347839,-64.8895112825383,-64.8598024276554,-64.9270631554447,-64.9394304031489,-64.8691865756608,-64.8396303112593,-64.6798326734477,-64.6267626156991,-64.5896761343386,-64.5053026856578,-64.5030901563902,-64.4337618751666,-64.3240664668155,-64.3278506455195,-64.3496554490041,-64.3869708078008,-64.4580309678366,-64.5115969334181,-64.4607241467232,-64.5282671589509,-64.5383379562229,-64.4933016475789,-64.5435335744648,-64.4886019380656,-64.5218508390342,-64.4727175516201,-64.4980547544022,-64.4169619459431,-64.3760226275539,-64.3608401317208,-64.3151782252874,-64.261795361348,-64.2943881310196,-64.3877032234388,-64.4603045232698,-64.4977801037759,-64.5227587272875,-64.5957872797565,-64.6702730428232,-64.6832582702245,-64.7792436602412,-64.8407899680941,-64.8294374401657,-64.8650209235032,-64.8329164391396,-64.8891145487171,-64.8392717278334,-64.8207094112248,-64.7657320020177,-64.7306673093416,-64.6516496963008,-64.6147692044112,-64.5501787695981,-64.4558031838549,-64.4206011632265,-64.3787005450819,-64.3502657931905,-64.3325427185381,-64.2876132320934,-64.233658168853,-64.2297061392949,-64.1787875762525,-64.1377948499268,-64.0871509482327,-64.0453418845432,-63.9954037040462,-63.9754719222698,-63.8621869004901,-63.8121342835392,-63.7776837704089,-63.7254644059152,-63.7099958138877,-63.6611181186575,-63.5953451444344,-63.5507895093629,-63.4675070876653,-63.6298490628446,-63.6606832465887,-63.7183576266113,-63.754375978979,-63.7942738796594,-63.8356175506359,-63.8550953816478,-63.9099659678729,-63.9502529606235,-64.047218712487,-64.1079257893022,-64.1049503194678,-64.0340809052337,-63.9735798290199,-63.9466938503577,-63.8871311957085,-63.849716661809,-63.8586926371835,-63.8078503699722,-63.8811154162425,-63.9242367380944,-63.9865803126894,-64.0305332345817,-64.1585010184899,-64.2169879381507,-64.2177737674703,-64.2474978777965,-64.3735583201705,-64.4217303067501,-64.4353945523309,-64.412277488176,-64.3539126344148,-64.2992709300833,-64.2692645293242,-64.2509616147394,-64.2185061849641,-64.2386630384416,-64.2274402016069,-64.0719989716786,-64.0259555885459,-64.0208820410446,-63.9385990529263,-63.9123577638488,-63.8834232946609,-63.8474812302034,-63.8042263988739,-63.7536282828502,-63.7485509244699,-63.6926084112097,-63.668053214804,-63.59677182239,-63.5987249498457,-63.6229635165634,-63.6846013643122,-63.69950537714,-63.7306142217732,-63.7455754542756,-63.7719159321597,-63.7511716108948,-63.7752232689966,-63.7612576645549,-63.7314610783645,-63.7164426257702,-63.7013020918872,-63.7854962419833,-63.80172776432,-63.8558201506704,-63.8833698881658,-63.8669781342643,-63.8840794166436,-63.8459858628668,-63.809132089884,-63.7597699320887,-63.7371373421418,-63.7709813159942,-63.7648854378462,-63.8118100057403,-63.8900799324993,-63.891937690632,-63.8577770846509,-63.9141277710893,-63.9180950584143,-63.863892040491,-63.8591580020407,-63.8995022255084,-64.0034793848987,-64.0127643603415,-64.0789111865798,-64.0762866772288,-64.1453174134015,-64.0908359210387,-64.0248569442376,-64.0504611844607,-63.9310650178375,-63.9306301370136,-63.8837170057185,-63.8742298591638,-63.9930156725229,-64.1527408078949,-64.1779864696819,-64.115356789627,-64.1278460951114,-64.0981906544982,-64.1486285575589,-64.0904925964073,-64.0873798071648,-64.0221485000058,-63.967110070016,-63.934894962486,-63.9129146909324,-63.9114956209979,-63.862938360196,-64.0359653335715,-64.0021366049391,-64.0346683328852,-63.948139584779,-63.8561787117736,-63.8382649089493,-63.8875812883651,-63.8396267480291,-63.8110546797922,-63.8378758078587,-63.714844242233,-63.6367230885522,-63.4957701024202,-63.4435507416609,-63.4912458742596,-63.4719931045149,-63.5630117355601,-63.6245389534729,-63.6546521588204,-63.6880879624293,-63.7123952012205,-63.7239766254987,-63.7925839251334,-63.854443028863,-63.825111830887,-63.8451008321898,-63.7529034510009,-63.7499699519271,-63.7803730783853,-63.7683987431947,-63.6665311196071,-63.6742062854305,-63.7258420011874,-63.6982083536969,-63.7251401012956,-63.7049298396236,-63.7511944686747,-63.7646221924345,-63.7159810141943,-63.6436925377339,-63.6877484565708,-63.6626515678451,-63.6914830326572,-63.7926830971974,-63.6653981431782,-63.4785658182653,-63.4349104419597,-63.3517996812253,-63.31979058076,-63.3248526764255,-63.442253726486,-63.5744100258537,-63.5908322972254,-63.5326810769971,-63.5225568719691,-63.6157613244451,-63.6794743736391,-63.6526990258506,-63.6012387806476,-63.6225057100939,-63.6090932363662,-63.5633130794592,-63.5474134272067,-63.4626356430593,-63.4028364867392,-63.4526106273769,-63.5629201684813,-63.5239187181577,-63.5734067625187,-63.5651288763635,-63.6022992676287,-63.5854116054373,-63.6160550497214,-63.5563932185416,-63.606133033301,-63.5575986571944,-63.57315879893,-63.6110082102986,-63.6113400895652,-63.5778775755566,-63.6122975785149,-63.5914426381722,-63.6307187394874,-63.6848645242737,-63.6964192394502,-63.7917141488076,-63.8298191529986,-63.7995762450407,-63.8377346393693,-63.8526577328227,-63.8171657959928,-63.9036640320132,-63.8908428366597,-63.9240078040898,-63.8029598702029,-63.7631039358102,-63.7238469023584,-63.7126240666109,-63.8133739946952,-63.8978656910279,-63.9744228135608,-64.003998153052,-64.054275846212,-64.0861133062265,-64.107330639399,-64.1027682613851,-64.1806872481591,-64.1776354893975,-64.2881930183983,-64.2946856269083,-64.3974764410938,-64.4927828088606,-64.4748003325526,-64.5341875295278,-64.5548174049916,-64.5306856300346,-64.5568849742904,-64.6799699700087,-64.6993638863108,-64.7348939699595,-64.7696687438163,-64.8010255486551,-64.7523347570947,-64.7762452830837,-64.7487413180343,-64.7611161959918,-64.7865373316958,-64.8007051192237,-64.8158647167987,-64.8301011643679,-64.8614884893333,-64.8484269716478,-64.9150392015033,-64.9719010732122,-65.0177232067469,-65.0858155478755,-65.126449692703,-65.1345139615497,-65.1803055860685,-65.2096177123126,-65.282737824286,-65.296402072744,-65.3357926288574,-65.3558426767419,-65.3845749740057,-65.4075470818046,-65.4113770315277,-65.4563370574734,-65.4872971347205,-65.4346237991929,-65.4291153795652,-65.455528337131,-65.5573883785897,-65.6469879818422,-65.693176328448,-65.757026724379,-65.8506393887497,-65.8498459389672,-66.0312195192697,-66.0408249287637,-66.152565033825,-66.1679764131557,-66.2355347004646,-66.2464218400219,-66.267974879262,-66.4447098034187,-66.4927444720506,-66.5164184813625,-66.5455932805906,-66.6180343787274,-66.6586380177088,-66.7002868867816,-66.8078460845082,-66.7998886246445,-66.8276825140918,-66.8237304841567,-66.707229626554,-66.6703643973629,-66.7552032654918,-66.7457275556463,-66.7760772849992,-66.7886734147212,-66.7549438654186,-66.6948623867038,-66.7536316054285,-66.7350387757905,-66.6716842872011,-66.6256103682699,-66.6501236176663,-66.7088089163354,-66.7659912250985,-66.681411756888,-66.6896515066777,-66.6026458886833,-66.6594314772648,-66.7373809955552,-66.7305374256644,-66.6795120367778,-66.6691131769862,-66.8756637728372,-66.9270477419303,-67.0330429202854,-67.0565109399601,-67.2727890072916,-67.263298037405,-67.2339782777394,-67.2728118973126,-67.4096298259433,-67.4637069754581,-67.4458694555937,-67.4019393959797,-67.3997573959891,-67.3577270563809,-67.2591781673628,-67.2213745177873,-67.2221450877678,-67.1774597282838,-67.0629043697597,-67.0715332096278,-67.1290054388443,-67.1385498087048,-67.1757736282322,-67.2295227176017,-67.2328338675507,-67.2803268470334,-67.2685012871489,-67.302703866785,-67.2622680772104,-67.2663192771611,-67.2268295375903,-67.2725753870724,-67.2897567769001,-67.3343048164471,-67.3296203664984,-67.3577957262242,-67.4290237455941,-67.4917450050771,-67.5268859947994,-67.474121095201,-67.5004043649765,-67.5816574143779,-67.6094741841792,-67.6474304239418,-67.6678085337998,-67.6966857936224,-67.7274932934466,-67.7669677732242,-67.7247009334463,-67.7480316233078,-67.6799545336906,-67.6808319136693,-67.615615844066,-67.6533584638227,-67.6444091838716,-67.7650375431734,-67.8102493329359,-67.7995071429835,-67.7580871631958,-67.7883606030368,-67.7910308830138,-67.8206329328629,-67.7762222330789,-67.7545089731944,-67.7336044333027,-67.6812667835988,-67.6692581236599,-67.5963058541186,-67.6091232340197,-67.5642623943181,-67.5690765442798,-67.4977874847941,-67.5360336345004,-67.6022262640443,-67.5572814943437,-67.6033096340281,-67.500000004752,-67.515968324624,-67.4651107850054,-67.4337387152576,-67.4406204252073,-67.4177932753971,-67.394729615554,-67.3769302457032,-67.3135528662836,-67.3186569262219,-67.2862930365339,-67.2574005168338,-67.2187728972324,-67.1890792875637,-67.180015567677,-67.1298904482604,-67.0359344594837,-66.9856338601821,-66.9958191000135,-66.9379196308912,-66.924446121067,-66.8900909516194,-66.9067077713352,-66.9463195907069,-66.9661026103893,-67.005729689812,-66.9969482499284,-67.0270462094924,-67.0188140995983,-66.9584198104674,-66.9533767805334,-66.9780578701157,-66.9596176203686,-67.0135269295754,-66.9951782298212,-66.9586944703534,-66.9908752498696,-66.9895248498682,-67.0684890787885,-67.0489273190804,-67.0752563587354,-67.1162033182035,-67.1344451980009,-67.2176971470662,-67.2629699766105,-67.3026733462111,-67.2880783163439,-67.342758185822,-67.346397405795,-67.3650054956259,-67.3619003356571,-67.3853759854537,-67.3634262155905,-67.3453674357437,-67.2445144767069,-67.2758331363866,-67.3214798059538,-67.3519363456676,-67.3245773358891,-67.2685318064201,-67.2274704068185,-67.1611328175253,-67.149497997677,-67.063148508741,-67.04766846892,-67.0688095186121,-67.0343017690429,-66.9495773402403,-66.8947677710221,-66.8511963017287,-66.8978958210089,-66.8559875616808,-66.8601837316492,-66.7649917732409,-66.7540664835174,-66.7871093829424,-66.7196731741925,-66.6866226348585,-66.6805038650099,-66.647415175703,-66.66509248537,-66.6453933858322,-66.6636047554573,-66.6327285961338,-66.6506042657662,-66.6258697663416,-66.5702819976082,-66.5203399888662,-66.5010910193401,-66.493011489609,-66.462219260435,-66.4753036701312,-66.4595337105737,-66.3938446223993,-66.3705826030999,-66.2581329565066,-66.2949447852593,-66.2835541055964,-66.3797378726313,-66.4165954815328,-66.3817672924623,-66.3445892535876,-66.3321457138999,-66.3086471846259,-66.3390884636054,-66.3039703646451,-66.2776947254975,-66.2802200553889,-66.2908325450158,-66.3093033044277,-66.3886566421133,-66.4402771205951,-66.371109032457,-66.368095422518,-66.399215721609,-66.3663864325403,-66.3555832128132,-66.3938675117071,-66.3931045717157,-66.3588180726587,-66.3729095722182,-66.3417129731273,-66.3440399430058,-66.3638458524018,-66.4198761208408,-66.448600790018,-66.4905166789147,-66.4527359198401,-66.4687652794067,-66.4328918703367,-66.4704208593288,-66.4317856003069,-66.4051285010548,-66.3568420623703,-66.3763656817692,-66.3332214630399,-66.3235702733067,-66.2679367350915,-66.3125686838668,-66.2834625447906,-66.2871017747085,-66.2666931453746,-66.2007904374942,-66.1723022784953,-66.1521072692704,-66.1039123810631,-66.075927762101,-66.0926285113032,-66.0663147323837,-66.0156326644783,-65.996192965275,-65.9703979864319,-65.9612808569507,-65.8977127499077,-65.8769913110185,-65.8053360447909,-65.7973633251938,-65.7530441776896,-65.7339401687314,-65.6682587228218,-65.6485596239661,-65.6512375437175,-65.6799088018723,-65.6773758419307,-65.65016942358,-65.6205444856734,-65.5793228684195,-65.5398941612302,-65.4993286744356,-65.4960785544672,-65.4561844573823,-65.4232025799685,-65.3997574617419,-65.3712158941406,-65.3712235242549,-65.3590851552344,-65.3493347959188,-65.3995056915132,-65.383392402713,-65.3625946744566,-65.3597565445294,-65.3315048970035,-65.3233032979017,-65.2835312715229,-65.2538681842506,-65.2588883235964,-65.2046128188236,-65.1696091624234,-65.1804200110343,-65.0911561104433,-65.0492173253391,-65.0100327599478,-65.0205231786518,-64.9508973369755,-64.9098511924443,-64.9386368988537,-64.9246751107545,-64.8478166010972,-64.8053819073103,-64.7731019519379,-64.7097474618303,-64.6916200243778,-64.6398774829917,-64.6241761950457,-64.601059129118,-64.5485078680857,-64.4553300965806,-64.4229967237369,-64.3910219403569,-64.3371202822259,-64.2740404577507,-64.3018877413454,-64.3024523217531,-64.352699500773,-64.3429720242746,-64.358772500837,-64.3396608652665,-64.3610384306179,-64.3228609493031,-64.2800981990457,-64.2371065696177,-64.2918627274289,-64.2515413872636,-64.2499239584483,-64.1663592196558,-64.1608050215379,-64.2006914813764,-64.2135088686554,-64.2339937834097,-64.217560068012,-64.2465593908652,-64.2098162403544,-64.2162249290497,-64.1915743651908,-64.1694567513008,-64.1827776680701,-64.1583559847255,-64.1652911031569,-64.1092303291426,-64.1093752893772,-64.1349719022984,-64.151947298851,-64.2257006399102,-64.2058108150516,-64.1946795282356,-64.1715395843195,-64.1545489292253,-64.172981535035,-64.1276629478516,-64.1404955847606,-64.0795367425075,-64.0535205101257,-64.0372699053089,-63.9374164273107,-63.9195102437984,-63.8886417243432,-63.8729404302812,-63.8189777496311,-63.7722248376361,-63.7214855676451,-63.717537359846,-63.6441235410356,-63.6765598977777,-63.6099629364384,-63.6198849627422,-63.5870633266234,-63.6165280320479,-63.5279927615108,-63.45004708931,-63.3838163522657,-63.4294210281164,-63.7666401172347,-63.9397281245466,-64.0326007142708,-64.0935137846486,-64.0206530854808,-64.0127185173589,-63.8732150825595,-63.7694820604479,-63.7428478504156,-63.8373912750847,-63.7706569891794,-63.801285177551,-63.7515682460437,-63.7001690343969,-63.702007712118,-63.6774944819016,-63.6463818238809,-63.8239749858576,-63.8401416697384,-63.8081096600316,-61.8065245299394,-60.4416945531855,-59.0344279718016,-57.1078383830923,-57.1084475780053,-57.1461586132602,-57.174932058677,-57.2080580047215,-57.2464977727547,-57.2439115647593,-57.2453725274865,-57.2525172780887,-57.4094842652117,-57.4198180102926,-57.4535200934314,-57.4658793952704,-57.4427209142651,-57.4409318242244,-57.4759651687197,-57.468469501806,-57.494340181348,-57.5071305171938,-57.5154616479798,-57.5526922337797,-57.5674433462314,-57.6097702636691,-57.6222669523056,-57.5851241182704,-57.6186545199585,-57.6925207422232,-57.6991620178096,-57.6793527115971,-57.7022556791993,-57.7038387739622,-57.6788072949671,-57.7181017133246,-57.6928870332866,-57.7195893253035,-57.7473254249608,-57.7328527779783,-57.755587909269,-57.7464137175906,-57.7905374115789,-57.7733334606081,-57.7736767431204,-57.8144779696833,-57.7875391340066,-58.006101817043,-58.0386406190431,-58.0282457729631,-58.0623065919128,-58.0458655937057,-58.0722627771635,-58.0775422397714,-58.147247025144,-58.0954367677705,-58.1633829369524,-58.2125078186542,-58.2111040831161,-58.2166924839063,-58.2357961874843,-58.2600496850181,-58.2448140529536,-58.2844672416944,-58.2811370862336,-58.3057491583506,-58.2620829868131,-58.2677286610572,-58.3352439638338,-58.3298424098654,-58.3003819404267,-58.2888426231006,-58.2696931246874,-58.2679765511388,-58.2560138152486,-58.2967580021808,-58.2721077640774,-58.2976391657914,-58.2860006565653,-58.3519481911179,-58.3676988605913,-58.3791351443368,-58.3999593101545,-58.4113917779271,-58.4291604186537,-58.4773279485295,-58.5806209066389,-58.5831767476232,-58.5898599874155,-58.613907574201,-58.6362461552601,-58.6155592791405,-58.6373714407989,-58.663028787762,-58.6639061805151,-58.6835516310416,-58.6651306536573,-58.6111304436793,-58.6333545891417,-58.5645001540441,-58.584691079207,-58.6112868250218,-58.6184964919414,-58.662796015303,-58.6681556102963,-58.7394628661463,-58.7850823102112,-58.8000166986248,-58.7990515689858,-58.8196125538949,-58.8396356950487,-58.8400171368642,-58.8716825856392,-58.8300189250731,-58.8244609555984,-58.8700155685036,-58.8916332041417,-58.916817571554,-58.8808033965603,-58.8897449372214,-58.9072389578078,-58.895016784927,-58.9066820109142,-58.9213684414635,-58.9242676063695,-58.8844616856455,-58.900582445287,-58.9055720104206,-58.9234551350998,-58.9572339081318,-58.9682811646745,-58.9725001663045,-59.0066833295268,-58.9829332545886,-59.0147894568502,-59.0466800293787,-59.0522379865523,-59.0237501017602,-59.0395465996769,-59.0106352918634,-58.9689029359379,-58.948540283008,-58.9855691432577,-58.9575771702099,-58.9571880636844,-58.9744608416675,-58.9574169408669,-58.9922371259078,-58.9522175374531,-58.9513897444772,-58.9800149527342,-58.9854088653984,-58.9716112297118,-58.9503788433741,-58.9522365699689,-58.9877929986246,-59.0173146274356,-59.0391573888837,-59.0616333584021,-59.036246788673,-59.0116803527277,-59.0758315216331,-59.0668518176799,-59.0917005246295,-59.10121050567,-59.1284832749244,-59.1027935768142,-59.0957631362832,-59.116800994236,-59.1107623952003,-59.1327488922383,-59.1410546229757,-59.2157609902616,-59.1879253828149,-59.2083452638827,-59.2013873243895,-59.2237794030003,-59.2317864019128,-59.2611249848798,-59.2633451183332,-59.2711232207513,-59.2569135735353,-59.325012978085,-59.3444563064771,-59.4289435769814,-59.4101296135931,-59.4372328307438,-59.4615933039678,-59.4761233593674,-59.4461972836848,-59.4205665045243,-59.4757571281174,-59.5053627881917,-59.4979089301215,-59.5142624214009,-59.5288992885477,-59.5597523443759,-59.5846506804729,-59.6054482665299,-59.5994554271428,-59.6215423678743,-59.6616765341001,-59.7124231188998,-59.736100793593,-59.7398735025948,-59.7861187861008,-59.806286976865,-59.79331327365,-59.8278055516829,-59.8723952761814,-59.8568123227462,-59.9011197772252,-59.8816763719825,-59.8272981779706,-59.8772322751633,-59.8515022888563,-59.8683402557031,-59.8311204722868,-59.8238535183036,-59.8477867861305,-59.8261194405253,-59.8079577712757,-59.9101986738039,-59.9072308667459,-59.9256366660581,-59.9503672105253,-59.9472315403431,-59.9721185018565,-59.9478304574717,-59.9600603141265,-59.975971325207,-59.97610865851,-59.9807892591904,-60.0216749629671,-59.9938468996115,-60.0061187271857,-60.0405614390905,-60.0238951009196,-60.0361173221263,-60.0688968571377,-60.0505520733275,-60.0616756797962,-60.0922312447718,-60.1003183580983,-60.0961184077662,-60.1633406553407,-60.1516753813624,-60.1927861722972,-60.1883382576009,-60.1366722560344,-60.17945006396,-60.1894521614551,-60.1472274798435,-60.1877851430911,-60.1806326308915,-60.1966733560938,-60.2500063754425,-60.2933411428786,-60.2438952665429,-60.2816453409455,-60.2905602677801,-60.2438952828438,-60.2888932568349,-60.3177856444552,-60.2922272867194,-60.3144516090853,-60.293924807503,-60.313894655409,-60.297228314738,-60.3144515916941,-60.3275397607985,-60.3416006719974,-60.3350050864346,-60.3772298003379,-60.3783398685966,-60.4200038285953,-60.4438951777515,-60.4472292094643,-60.4994483938912,-60.5072265383664,-60.5489629703987,-60.5938923169409,-60.6004573808793,-60.6311160043958,-60.6364985222494,-60.6416712293487,-60.6756829502735,-60.6933373117697,-60.6794480519722,-60.6972969575622,-60.7111175636567,-60.7100036652394,-60.7392050787165,-60.7283370479355,-60.7511145345586,-60.8077168150449,-60.8366702903044,-60.8678515222301,-60.8361171662979,-60.7816701599382,-60.7950024883491,-60.8538936027638,-60.8405612729101,-60.8894502784496,-60.8972246082956,-60.9138909670637,-60.9235802705534,-60.9443779343837,-60.9588928285769,-60.937225411872,-60.9488945411672,-60.9194489768908,-61.0000037065655,-61.0288923325243,-60.998336690642,-60.9583358839944,-60.9550056607907,-60.9711150817385,-61.0396039679677,-61.0530659993597,-61.0856663206486,-61.1355090366048,-61.1685976328057,-61.1291537619362,-61.0873867458641,-61.0992237218372,-61.0655438364472,-61.0626370382304,-61.107879235472,-61.1233249033761,-61.1022563865804,-61.1202502698177,-61.1761316339856,-61.1805070787845,-61.1924851919767,-61.2272026688007,-61.2135079427161,-61.2636367645765,-61.2663757053515,-61.2915030605443,-61.325041805115,-61.7308904716608,-61.8125172322618,-61.7606450647595,-61.6947081274659,-61.6332267653899,-61.5832123582812,-61.5838045808316,-61.6019386734116,-61.5899846052464,-61.6375373684279,-61.7936383238172,-61.810537407,-61.824174930786,-61.8047085625236,-61.8405781058875,-61.824777661856,-61.864393230117,-61.8399677590706,-61.8736514886187,-61.8889407643186,-61.8999270796429,-61.9295977550048,-61.9211024393684,-61.9338358794976,-61.964673846188,-62.0011117834268,-62.0277802983472,-62.0514046860215,-62.1572547721304,-62.2127776146895,-62.2889417794389,-62.3149961300321,-62.3426374015218,-62.3650029466282,-62.386666584735,-62.3994458104232,-62.4100010715329,-62.4434063354856,-62.4892551417778,-62.520749250902,-62.5494471954894,-62.5685053971337,-62.6394472647956,-62.6138888163852,-62.654999771638,-62.7202272272555,-62.7388887131499,-62.7670297104483,-62.7033357630874,-62.7691316094216,-62.7715463084895,-62.7888879039757,-62.8320702509815,-62.8766678384728,-62.8772247886548,-62.9650123572395,-63.0171783046255,-63.0577780983586,-63.0327765938113,-63.0544440596575,-63.1112143449828,-63.1584555378679,-63.192223218405,-63.1549841585208,-63.2028280610855,-63.2161108341194,-63.221111900724,-63.3589864301875,-63.4004864994301,-63.4479641976331,-63.4659390505766,-63.4925351090707,-63.5495648048616,-63.6072239309884,-63.6448787969299,-63.687221920645,-63.7155574810018,-63.7996753415181,-63.7900012752072,-63.8533328538756,-63.9332850691676,-63.9844439539788,-63.9961131108305,-64.0162552845247,-64.021199991448,-64.0299990612898,-64.0243819676984,-64.0894472850334,-64.1640245955978,-64.3398439559776,-64.4577791128682,-64.5444413867934,-64.6972199919635,-64.7438889849687,-64.7838898994039,-64.8750001171405,-64.927780260664,-64.9261456868747,-64.9583359869653,-65.1222229893382,-65.2286835491975,-65.2386627983852,-65.3157654517218,-65.3491593090544,-65.689025926056,-65.738334703383,-65.746856732975,-65.7572250823888,-65.7969818503809,-65.8017349601764,-65.8194427892557,-65.9499969834513,-66.0065536810832,-66.0566635390419,-66.1116638470274,-66.2366638430069,-66.3633346793995,-66.4100341982619,-66.3935012986867,-66.4083328383606,-66.4555587972276,-66.5400619652709,-66.5144424557602,-66.4238891778438,-66.4517364671341,-66.5616684147504,-66.7066650519854,-66.735748301431,-66.8094482502649,-66.8150024501715,-66.8922195490818,-66.9563522382258,-66.981887827874,-66.9626693781032,-67.0433349671084,-67.0580520669468,-67.0611114569284,-67.0894470266362,-67.1483306960331,-67.1374969561311,-67.1660808531105,-67.1655578657945,-67.2344436651557,-67.2255554251729,-67.279129034714,-67.3027572645342,-67.3044433645091,-67.3799972539477,-67.5888900827618,-67.6025009226908,-67.6272201525777,-67.6055526726765,-67.6444473324989,-67.6716690123755,-67.7897186319164,-67.7669448919964,-67.817779541807,-67.892776491571,-68.0274963412017,-68.0497207611514,-68.0778656010798,-68.1271286009772,-68.1444473309315,-68.2646942107139,-68.3241119406251,-68.231170650766,-68.1884994508414,-68.190551760829,-68.3119430506309,-68.3977813705104,-68.4645614604328,-68.3930587805217,-68.376686100547,-68.4295806904758,-68.5249252303735,-68.552078250346,-68.5611648603386,-68.6002807603036,-68.627464290279,-68.6148605302883,-68.6366653402709,-68.6408309902623,-68.6927795402253,-68.6561126702494,-68.6186370802786,-68.6374969502623,-68.7841644301665,-68.8496246301299,-68.8700027501205,-68.8758316001169,-68.939170840091,-68.9241638200965,-68.951110840086,-68.9288864100941,-68.9794464100738,-68.9891662600714,-69.0100021400634,-69.0013885500669,-69.0620193500483,-69.0430755600527,-69.0828704800401,-69.0942840600332,-69.0930938700349,-69.1451034500205,-69.2153091400077,-69.2284393300053,-69.2806167599956,-69.2822799699942,-69.3209152199879,-69.4032516499783,-69.3910369899789,-69.4247207599754,-69.4980544999702,-69.5155868499688,-69.5529785199671,-69.5511474599674,-69.5711746199662,-69.5782241799664,-69.6118850699646,-69.6805267299628,-69.7166137699626,-69.7278747599622,-69.7066879299621,-69.7562866199616,-69.7766647299606,-69.8147201499608,-69.7958297699605,-69.8025588999601,-69.8669433599605,-69.9054260299604,-69.9575729399615,-69.991561889962,-70.1263885499658,-70.1563873299668,-70.139640809966,-70.1991653399678,-70.2030563399676,-70.2311096199684,-70.4527740499758,-70.4948348999772,-70.4977798499771,-70.5505523699787,-70.5722198499792,-70.7433319099838,-70.8055572499854,-70.9030151399877,-71.0116653399899,-71.0566635099908,-71.1399993899922,-71.1536102299924,-71.192222599993,-71.2055587799932,-71.2830581699942,-71.4935302699965,-71.6373596199976,-71.7207412699981,-71.8768920899988,-72.0076217699992,-72.1947097799996,-72.2423171999997,-72.2592544599997,-72.4843978899999,-72.5692520099999,-72.6557388299999,-72.70494843,-72.84984589,-72.95257568,-73.069664,-73.17274475,-73.20175171,-73.25400543,-73.46996307,-73.52333832,-73.62114716,-73.68586731,-73.77855682,-73.75531769,-73.67562103,-73.61927032,-73.478508,-73.47283936,-73.52846527,-73.52641296,-73.56504059,-73.62784576,-73.78321838,-73.81227112,-73.95320892,-73.97245789,-73.92536926,-73.93952942,-73.93131256,-73.87625885,-73.85512543,-73.87081909,-73.85478973,-73.82047272,-73.87947845,-73.94051361,-74.02832794,-73.97669983,-73.89045715,-73.89025879,-74.05252838,-74.14891815,-74.19514465,-74.22605133,-74.32318115,-74.4722061200001,-74.3812713600001,-74.4361496000001,-74.4723663300001,-74.5351104700001,-74.6358947800002,-74.7240295400003,-74.9367752100007,-75.035469060001,-75.1278686500013,-75.2364120500018,-75.3425140400025,-75.6775665300059,-75.7046890300063,-75.7563552900071,-75.8178253200082,-75.8963699300096,-75.9871597300115,-76.089874270014,-76.2010040300173,-76.2334823600183,-76.2450866700188,-76.3659591700231,-76.5010681200285,-76.6079788200335,-76.660011290036,-76.6712112400363,-76.7119445800382,-76.689331050037,-76.692893980037,-76.7679824800408,-76.7813873300412,-76.76473999004,-76.7810363800407,-76.8948440600467,-76.9275665300485,-76.9023513800474,-76.9274749800488,-76.9133682300483,-76.9353485100497,-76.9876861600527,-77.0815582300577,-77.1903305100635,-77.278404240067,-77.2791366600662,-77.5660781900727,-77.6757278400715,-77.6946563700716,-77.7955856300675,-78.0000000000457,-78.0555496200369,-78.1299438500199,-78.2604904199825,-78.3123626699649,-78.3883895899287,-78.5026626598648,-78.5970611597952,-78.711494449692,-78.7271041896684,-78.8927230794488,-78.9898681592749,-79.0152511592129,-79.0943221990383,-79.1708145088248,-79.2138442987088,-79.2768325785089,-79.3296661383205,-79.3467941282462,-79.4349899278896,-79.4268417378652,-79.5868835370503,-79.5532302871923,-79.5104827873898,-79.5189208960302,-79.51565551591,-79.4982910160211,-79.5400009157111,-79.4727783161518,-79.3555526668101,-79.3105697570429,-79.2788925172051,-79.2272186274428,-79.254394527347,-79.1761093076899,-79.0955810479969,-78.9893798783617,-78.9499969485067,-78.9349975585514,-78.9085082986228,-78.8732910187027,-78.846107478768,-78.8833312986751,-78.8922119086475,-78.7755584689117,-78.711112979042,-78.7083129890458,-78.7766647289073,-78.8255538887952,-78.8261108387847,-78.8051452588296,-78.839996338748,-78.8811035186394,-78.9266662585153,-78.9411010684669,-78.9916686983138,-78.9988861082849,-79.0416641181411,-78.9748840283492,-78.9144287085284,-78.8926315285846,-78.8444213887078,-78.8193283087618,-78.7727813688684,-78.779418948851,-78.8333358787255,-78.8612060486538,-78.838722228708,-78.8968505885571,-78.8560943586598,-78.7214736889647,-78.6978759790098,-78.7249755889532,-78.6772232090479,-78.6927185090168,-78.6500244090962,-78.6838912990328,-78.7055053689879,-78.6422195391082,-78.6306304891271,-78.657775879077,-78.5755538892168,-78.5266647292823,-78.5388870192651,-78.5610961892283,-78.4883346593361,-78.5026855493147,-78.4777755693496,-78.5478210392473,-78.5638885492187,-78.5054931593059,-78.5033340493052,-78.4855422993306,-78.5433349592438,-78.5100021392939,-78.5250091592703,-78.5011444093036,-78.5466918892339,-78.5283203092608,-78.5077743492919,-78.4944610593085,-78.5459670992301,-78.5194473292702,-78.5566635092108,-78.5594482392047,-78.5871963491586,-78.5805053691687,-78.6416702290615,-78.629394529083,-78.6549987790348,-78.6372222890673,-78.6799926789842,-78.7172241189096,-78.7044448889355,-78.7272186288868,-78.6893920889657,-78.7410888688545,-78.7004775989401,-78.7694396987907,-78.757202148816,-78.7293167088785,-78.7027664189326,-78.7349853488631,-78.708312988919,-78.7366638188577,-78.7027740489295,-78.7383346588525,-78.7133331289042,-78.7322235088625,-78.7327804588636,-78.7683105487839,-78.7683029187815,-78.8132705686728,-78.7438888488359,-78.7094421389084,-78.7371826188467,-78.8094482386769,-78.8265991186309,-78.7776946987522,-78.7661132787775,-78.7927780187125,-78.7611083987878,-78.7905883787161,-78.8116683986657,-78.8533325185542,-78.8727493285017,-78.8199996886375,-78.7933349587076,-78.7532958988009,-78.7661132787699,-78.7388915988341,-78.7072219789017,-78.738891598833,-78.7227783188672,-78.7511138888027,-78.7216644288654,-78.7716064487539,-78.7921752887028,-78.7238769488591,-78.7422256488169,-78.6978073089129,-78.6927795389247,-78.6888885489312,-78.7227783188578,-78.8110961886506,-78.8405532785712,-78.8800048784641,-78.8400268585721,-78.8733367884814,-78.8705367984873,-78.8927764884242,-78.8699951184866,-78.9072189283787,-78.9116668683637,-78.8261108385983,-78.7744445787287,-78.7783355687178,-78.8405609085579,-78.9100036583627,-78.8010864286593,-78.8561096185135,-78.8027801486533,-78.8450012185428,-78.8466873185352,-78.8744049084599,-78.8600006085025,-78.9299926782979,-78.9111099183533,-78.9533081082227,-78.9254760683078,-78.9644470181859,-78.9921874980895,-78.9494018582294,-78.9844436581124,-78.9404449482568,-78.9205551083168,-78.9427795382482,-78.9210815383141,-78.9550170882048,-78.9483337382247,-78.9916686980833,-78.9566650381964,-78.9738922081386,-78.9466628982238,-78.973335268136,-78.9205551082997,-78.9738464381268,-78.8966674783667,-78.9405517582301,-78.9288940382654,-78.8927764883759,-78.8983306883564,-78.9511108381919,-78.9033203083382,-78.9849548280722,-78.9744035624351,-79.0125503479757,-78.9829025280765],"lat":[53.292171477429,53.2856000388442,53.3052787775464,53.3261108375017,53.3236122074205,53.3427772475275,53.3497238174425,53.3567161575723,53.3666648875708,53.3636093074961,53.3863906875491,53.3866653375025,53.3967285175714,53.4011115975132,53.4047241175729,53.3974990774435,53.4263877874095,53.4077758771937,53.4421997074185,53.4542846672727,53.429077147179,53.4440917971424,53.4549064572519,53.4530639571052,53.469722747066,53.4724273671337,53.4902763369507,53.4805297871244,53.4917831370196,53.5145034771367,53.5158691370702,53.531944267394,53.5587158172702,53.5561103774022,53.5374755874896,53.5574989276878,53.5724983174643,53.5981063775163,53.6044921873735,53.6168594373982,53.5861129773142,53.5852775571917,53.6280670172446,53.6279296873346,53.6084976172813,53.635223387402,53.6444435072111,53.652278897323,53.663333887235,53.6638793873398,53.6836128174232,53.6810111972869,53.6947212172335,53.6831970171858,53.6872215270598,53.6981201170911,53.7007751470139,53.6933326669227,53.7058715769213,53.7135009770304,53.7000007571234,53.7102241471685,53.6941680873528,53.7011108374421,53.7133140571444,53.726318357349,53.7411117570833,53.7391662572245,53.7599601671541,53.7580566372216,53.7866668672987,53.7857131971518,53.8188018772759,53.8169441172278,53.8574981671639,53.8808326672227,53.9005927970831,53.902568817266,53.8847465474486,53.8989257775233,53.9083328173571,53.9288940373984,53.9157638473633,53.9454193069992,53.9349174472765,53.9494438170727,53.9497299173895,53.9560394272847,53.9692192072897,53.9680557270868,53.9769287072559,53.9940795871814,53.9959106375299,54.0183105473038,54.0341682372817,54.0236816371257,54.0436096171864,54.0427246070578,54.0544433571886,54.0831146168676,54.1016845669028,54.0847206070015,54.0897216772374,54.1097526572344,54.1375122070569,54.1647224372278,54.1650009168498,54.158332816729,54.1425018267567,54.1413879365983,54.1519164965616,54.1680564867255,54.1869430466346,54.1702766364062,54.1791648864329,54.1725006063261,54.1833343463057,54.175834656088,54.2055549562309,54.2197265565044,54.2258911064445,54.2361144965129,54.2496948161387,54.2586097661561,54.250831596005,54.2641677859855,54.2725639262123,54.26751708595,54.2836112960439,54.2994461059981,54.3036117557123,54.3199996858432,54.3375244057258,54.3386115958554,54.3508338857309,54.3672218257456,54.365001675613,54.3938903757375,54.4036102256435,54.3991661058006,54.4130859358566,54.4133338856439,54.4241676257669,54.4338874756852,54.451946255759,54.4613876255209,54.4647216757011,54.4874992355271,54.4960937456767,54.4999999954837,54.505279535634,54.5144462556225,54.506389615545,54.5253295856158,54.5313873255268,54.5400123556117,54.5550003052817,54.5544433554767,54.5897216756501,54.5875244054537,54.6197204554973,54.6036109853249,54.6236114453912,54.6169433552263,54.6294441153515,54.6116943349421,54.6369438150107,54.6355743345923,54.7211303653715,54.7336120557284,54.7555541958659,54.7416992158311,54.7383346559127,54.7639160161306,54.7583351063083,54.7972564667181,54.8380012473922,54.8328552174719,54.8549995375849,54.871704097576,54.883888237716,54.895324707704,54.9119224478501,54.9075317380366,54.9338951081269,54.9603271483949,54.9920844984537,55.0423011788302,55.1229247990003,55.2180786093062,55.2578735393673,55.2621612494145,55.3449211094888,55.3828735395377,55.3970756495843,55.4927558896618,55.5247306797019,55.5297584496954,55.5614433297173,55.5870208697456,55.6554031397747,55.6627883897894,55.7211303698029,55.670104979761,55.7486114498016,55.7483291598087,55.8236122098306,55.8219451898391,55.8277244598346,55.9136123698609,55.9302787798732,56.0009155298809,56.014999389894,56.047779079902,56.1463890099109,56.165000919908,56.1580810499212,56.1421947051217,56.1511230499262,56.1441650399314,56.141577117182,56.1322212199276,56.1380538899357,56.1045341499527,56.1163902299564,56.0977783199686,56.1077270499696,56.101665499973,56.1204872099736,56.1311035199765,56.1172218299795,56.1260986299823,56.1680564899778,56.1811103799802,56.1969451899764,56.2089080799773,56.1994438199756,56.2205543499755,56.2341651899713,56.2569427499704,56.2863883999619,56.286594389968,56.3008346599624,56.3233337399625,56.3486099199518,56.3563880899577,56.3694457999537,56.3705558799585,56.3820533799537,56.3811111499614,56.4025268599629,56.3822212199679,56.4108314499681,56.4135627699698,56.4183349599676,56.4002761799657,56.4061126699612,56.4094848599642,56.4161109899627,56.4191665599536,56.4458351099575,56.4477767899515,56.472473139951,56.4847297699467,56.4891662599529,56.5311279299453,56.5475006099485,56.5338745099417,56.5080566399469,56.4886093099437,56.4705543499472,56.4099998499383,56.3833007799443,56.3680572499396,56.3510742199399,56.336120609947,56.3130569499479,56.2896728499418,56.2894897499346,56.2574996899386,56.2397384599311,56.2127761799328,56.1771240199196,56.1819496199158,56.1716918899192,56.1817016599334,56.1674652099362,56.1577758799293,56.1683349599088,56.1902198799105,56.1942977899065,56.1961059599144,56.2374877899137,56.3032836899261,56.3491210899245,56.3718757599287,56.4536743199303,56.4757080099257,56.518127439925,56.646728519929,56.9110526999277,56.9266662599239,57.021667479923,57.0938720699267,57.3124999999151,57.3392906199085,57.4046173099075,57.4080810499033,57.4481964099008,57.459472659892,57.4692344698947,57.6259155298698,57.6327247598754,57.6608886698713,57.6666870098756,57.8166465798376,57.8427772498376,57.8682861298218,57.9403076198095,57.9558982797903,57.9636115997943,57.9769439697822,57.9883956897903,57.9996108997731,58.0194892897783,58.0294799797596,58.0877380397204,58.0981101996923,58.1331176796819,58.1608314496252,58.1646728496673,58.1949081396317,58.2125244096418,58.2288818396178,58.2387466396228,58.2233314496111,58.2305297895803,58.2404670695974,58.2447395295651,58.2747268695356,58.2863883994709,58.3127937294387,58.3400001493103,58.3756256092209,58.3934097292088,58.3904838592689,58.4251098592266,58.4272117591747,58.4507265495865,58.4459037791679,58.4584045391291,58.4464721691288,58.4495162990922,58.4581260691123,58.4945030190163,58.5073242190312,58.5073242190002,58.5578651390575,58.526458739014,58.523502348946,58.5121803289939,58.5128784189309,58.5567398089147,58.5529174788866,58.593887328845,58.580268858839,58.5900726288146,58.6184043889146,58.6119346587692,58.6447944586757,58.5981445287667,58.5761833187358,58.5645141587623,58.5706787088122,58.5334052988798,58.5405731187687,58.5761833186833,58.5800971987146,58.6298828086663,58.6433105485195,58.6665039085482,58.6691284184992,58.6787109385176,58.6542396486071,58.663959498613,58.6478500386618,58.656677248678,58.6742362986317,58.6660766585986,58.6922950685691,58.6861839286732,58.729812618658,58.7566680885591,58.7739601085804,58.738960268664,58.761180878649,58.7411956787519,58.7094726587633,58.709712978698,58.7062263487698,58.6885337788045,58.7118720988243,58.7020797688919,58.7125015288393,58.7294959988449,58.7166747988388,58.7259063688046,58.7397918688108,58.7320175187664,58.7606277487212,58.7613906886731,58.776039118671,58.7641601586518,58.7753486586035,58.7711830086584,58.7963905286129,58.8339233386416,58.8263893085997,58.8416671785586,58.9187011685433,58.9167365985892,58.9471015886103,58.9302978486629,58.8627777085952,58.8651428186575,58.8939628586497,58.8950729386989,58.8353500386453,58.8573684686744,58.8637084987369,58.8695144687017,58.9156265287179,58.8999900787705,58.9097938488279,58.8984756488366,58.923095698889,58.9322929389709,58.9288825989131,58.9464721688899,58.9822997989811,59.0500144989401,59.0641670189771,59.046119688994,59.0383300790623,59.1055297890298,59.1016654990631,59.0683326690744,59.0911102291078,59.0847167992207,59.0977783191628,59.1175003091894,59.0958900491556,59.098331449125,59.109497069153,59.1195068391194,59.1202773991754,59.1199989291225,59.1494445790957,59.1474990791738,59.175556179147,59.1798779771568,59.1988906891429,59.1952781691687,59.2141685491686,59.2033576991948,59.2297210692468,59.2174987792564,59.2163886992794,59.2288818392637,59.2119445793231,59.1921653693331,59.2249984693218,59.2308349593665,59.2305870093002,59.2508315992834,59.2468872093077,59.2622642493081,59.2544441193483,59.2697219793579,59.2622222894052,59.2739257794032,59.2772216794272,59.3074989293725,59.3158340494108,59.2813873294429,59.2996826194364,59.3022346494547,59.3195190394342,59.2999992394679,59.3172225994857,59.3230552694593,59.3411102294801,59.3630981394312,59.3572235094772,59.3691673295047,59.3772239694868,59.386718749536,59.3761901895681,59.4058837895143,59.3936767595167,59.4011955294861,59.3861122094934,59.4188880894528,59.396110529367,59.3855552693738,59.4052772493451,59.4236106893728,59.437499999356,59.4236106894162,59.435276029437,59.4697227493823,59.470832819402,59.4897232093827,59.4900016794047,59.5067138693997,59.525833129492,59.5483322094995,59.5440063495261,59.5819435094982,59.6027793895186,59.6027793894971,59.608333589513,59.622776029506,59.6205558794787,59.6408653294851,59.6391677894673,59.6513900794789,59.6785545294532,59.6855544994991,59.707275389467,59.696945189512,59.7091674795137,59.712223049546,59.7366943395432,59.7327766395712,59.7144775395647,59.6933326695937,59.6844444295547,59.7067909195257,59.685001369525,59.6677780195481,59.6827773995582,59.6716918895644,59.6781120295879,59.6652831995701,59.6522216795884,59.6619262696075,59.6836280795942,59.6975097696057,59.6586112996118,59.6552734396417,59.6118850696691,59.6691932696646,59.6759033196402,59.6630859396341,59.6874999996305,59.6852760296203,59.6972351096255,59.6874999996413,59.7058715796523,59.7075195296207,59.7555541996217,59.7527770996463,59.7322235096524,59.7509002696734,59.7722206096659,59.7836914096843,59.767776489692,59.7900543196967,59.7830810497386,59.7991676297562,59.8061103797252,59.8257484397493,59.8280563397295,59.9030761697059,59.8855552696824,59.9256362896742,59.9214897197101,59.9260215796955,59.955421449704,59.9568939197385,59.9827194197327,59.985465999755,59.9999999997396,59.9999999998009,60.0046844497367,60.0187492397637,60.0109405497726,60.0442695597926,60.054164889785,60.0187492397288,60.0583343497076,60.0291671796801,60.0458335896639,60.0312499996553,60.0609397895867,60.0651016196116,60.109893799607,60.1234397896791,60.185939789628,60.2000007596313,60.1895828196478,60.2036437996761,60.2088546796591,60.2244758596507,60.234893799661,60.260940549628,60.2630195596497,60.2994804396348,60.2770843496291,60.2942695596088,60.3119811996372,60.3083343496181,60.3291664096114,60.3296852095838,60.3661460895839,60.3526039095613,60.3671874995376,60.3786430395479,60.3880195595072,60.405731199509,60.4130210895296,60.4291648895288,60.4317741395556,60.4536437995487,60.4671897895794,60.4807281495739,60.4963569596243,60.536975859629,60.5328102097186,60.5624999996618,60.5624999996234,60.541667939576,60.5807304395398,60.5942687994943,60.6390647894638,60.6578102094688,60.6942710895606,60.7328147895748,60.75051879961,60.7588539095964,60.7708320596059,60.7567710895453,60.783332819588,60.772396089547,60.8348960896677,60.7895851095112,60.7645835894587,60.7755241394325,60.7979164094595,60.7833328193613,60.7916679392575,60.8124999992642,60.7838516191172,60.8598937991544,61.0536460894522,61.0380210894711,61.1453094494879,61.1744804395187,61.1734352095386,61.1890602095329,61.1791648895576,61.1880187995653,61.216148379562,61.2296905495338,61.2640647895459,61.3119811995288,61.318748469558,61.3354148895478,61.327606199525,61.3765602095363,61.4578094495027,61.4619789095301,61.4171905495552,61.3994789095921,61.3812484695822,61.3942718496249,61.4255218495721,61.4557304395753,61.4562492396218,61.4807281496549,61.5151061995752,61.5390624995757,61.5333328196081,61.5562515296281,61.5458335896647,61.5192718496861,61.5494804396856,61.5723991396092,61.5755195596501,61.6182289096428,61.6624984697255,61.6562499996845,61.6375007596753,61.6453094496519,61.5937499996344,61.5916671795651,61.6244811995617,61.6421852095322,61.6869811995271,61.6833343494015,61.7255210893533,61.7640647893589,61.8609352092953,61.9286460893043,62.0026016192436,62.03697585926,62.1484374992168,62.2104148892534,62.2973937992554,62.308856959307,62.3182258592681,62.3296852092965,62.3515624992926,62.3651046793474,62.379165649334,62.3734397893085,62.3890647893128,62.4015655493903,62.3682289093502,62.349998469358,62.3895835894216,62.3812484694605,62.4244804394468,62.4104156494794,62.4265632594698,62.4291648895143,62.4578094495466,62.4692687995892,62.5125007596141,62.5250015296476,62.5130195596649,62.5765647896979,62.5791664097334,62.5250015297418,62.5562515297559,62.5541648897793,62.5348930398238,62.5041656498254,62.5286483798299,62.5333328198423,62.49166488985,62.5291671798518,62.5187492398559,62.5291671798574,62.5354156498822,62.5088539098751,62.5286483798891,62.4833335898885,62.5223960898933,62.5125007598959,62.5208320599052,62.5119781499121,62.4958343499114,62.4973983799195,62.4749984699216,62.485416409925,62.4744758599303,62.4661483799264,62.4291648899265,62.4583320599314,62.4249992399368,62.4562492399404,62.4333343499428,62.4458351099449,62.4208335899536,62.4291648899566,62.4062499999612,62.4187507599638,62.3791656499722,62.3770828199764,62.3588561999776,62.3333320599763,62.3541679399807,62.308334349981,62.3432311999825,62.3499984699843,62.3062515299889,62.2958335899924,62.2723960899942,62.2041664099904,62.1932258599881,62.1723937999866,62.1604156499869,62.1624984699887,62.1484374999897,62.1848983799894,62.2067718499927,62.230728149993,62.2505187999948,62.3020820599958,62.2630195599962,62.2583351099966,62.3057289099964,62.3166656499971,62.2744789099987,62.2229156499986,62.2421874999988,62.2687492399988,62.2541656499994,62.2182311999995,62.1942710899995,62.2020835899996,62.1703147899997,62.1380195599997,62.1583328199997,62.1187515299998,62.1062507599999,62.1442718499998,62.2088508599997,62.2515602099997,62.24583435,62.2791671799999,62.2734375,62.31406021,62.3312492399999,62.34166718,62.32291794,62.35416794,62.36302185,62.40156555,62.46250153,62.47916794,62.42656326,62.4067688,62.4057312,62.37760162,62.38750076,62.35625076,62.375,62.32708359,62.3119812,62.26927185,62.20156479,62.1796875,62.19374847,62.16510391,62.14166641,62.12708282,62.12031555,62.1088562,62.10833359,62.125,62.11249924,62.13072586,62.14166641,62.1098937999999,62.06927109,62.0588569599999,62.0494804399999,62.0390624999999,62.0244789099999,61.9692687999999,61.9598960899999,61.91093445,61.84635162,61.8442688,61.87239838,61.88124847,61.8359374999999,61.81093979,61.89374924,61.9500007599999,61.9249992399999,61.9244804399999,61.9000015299999,61.8895835899997,61.8578147899996,61.8338508599996,61.7942695599994,61.7859382599997,61.7765655499995,61.7598991399996,61.7416648899996,61.7437515299995,61.7666664099994,61.7557258599993,61.7458343499994,61.7453155499992,61.664585109999,61.6020851099993,61.5999984699995,61.6187515299996,61.6083335899997,61.5671882599997,61.6062507599996,61.5833320599994,61.596351619999,61.6546897899987,61.7062492399989,61.7015647899986,61.6895828199987,61.6937484699982,61.6729164099981,61.6833343499979,61.6687507599979,61.6562499999968,61.6203155499958,61.5854148899955,61.6036491399953,61.5651016199956,61.5661430399964,61.5453147899961,61.5354156499963,61.5541648899968,61.5348930399983,61.5348930399977,61.5182304399979,61.4880218499977,61.4921874999981,61.4671897899974,61.4666671799978,61.4307289099985,61.4062499999972,61.4098930399959,61.4078102099967,61.3963508599964,61.3994789099968,61.3749999999968,61.3749999999983,61.3619804399979,61.3458328199982,61.3520851099972,61.3276061999971,61.3265647899968,61.3286437999977,61.3109397899972,61.2859344499977,61.2729148899966,61.2494811999959,61.1994781499966,61.1895828199965,61.2098960899963,61.2182311999956,61.1776008599955,61.165103909996,61.1671905499948,61.2203102099952,61.224998469994,61.169269559994,61.1624984699938,61.1770820599933,61.1661491399928,61.1479148899932,61.1453094499927,61.1229171799931,61.1145820599927,61.1411437999925,61.1583328199916,61.1458320599905,61.1578102099888,61.142185209989,61.1348991399881,61.0958328199875,61.1203155499877,61.1416664099866,61.1187515299845,61.1333351099827,61.12499999998,61.0994758599762,61.0776061999771,61.0770835899717,61.0536460899672,61.0166664099646,61.0729179399672,61.0833320599655,61.068748469965,61.0770835899634,61.060939789963,61.0687484699612,61.0515632599571,61.0187492399593,61.0374984699547,61.0145835899518,61.0145835899464,61.0411491399484,61.0401039099438,61.0666656499412,61.0276069599375,61.0062484699305,61.0661430399334,61.0520820599249,61.0875015299308,61.0869789099263,61.0583343499209,61.0807304399219,61.083854679916,61.0979156499153,61.0869789099038,61.0604171799033,61.0895843498847,61.0609397898866,61.0687484698803,61.0588569598786,61.0026016198888,60.9598960898739,60.8838539098711,60.8895835898837,60.8796844498856,60.8812484698724,60.8380241398673,60.8437499998514,60.7999992398268,60.8557281498054,60.8494758598191,60.8229179398252,60.8432311998341,60.86302184981,60.8890647898066,60.9145851097822,60.9119758597719,60.8749999997751,60.8817710897355,60.9161491397342,60.9458351097551,61.0223960897509,61.0255241397347,61.0515632597414,61.0770835897265,61.0703124996828,61.0223960896554,61.0265655496685,60.9609374996507,60.9494781496335,60.9317741396437,60.9140624996076,60.8859405496094,60.8875007596243,60.8067741396009,60.7958335896052,60.822917939622,60.814060209634,60.7859344496274,60.7578124996877,60.7369804397074,60.7369804396966,60.7213516196993,60.7333335897292,60.7088546797265,60.6953124997577,60.6619758597464,60.638019559753,60.6312484697375,60.5963516197452,60.5953102097287,60.5817718497276,60.5723991397425,60.5583343497392,60.5578155497562,60.5442695597475,60.5437507597858,60.522396089794,60.4838561997836,60.4578094497611,60.4151039097771,60.4036483797598,60.3817710897608,60.3557281497405,60.3557281497517,60.3395843497501,60.3411483797635,60.3067741397776,60.2848930397397,60.2609405497449,60.2598991397291,60.231769559724,60.2244758597052,60.1963539097113,60.1895828197325,60.1791648897063,60.1411437997256,60.1130218497122,60.091667179734,60.0713539097154,60.0645828197562,60.0395851097583,60.0723991397963,60.0213508597993,60.0140647897797,59.9928207397818,60.0333328199147,60.010940549907,59.9999999999496,60.0229148899527,60.0312499999676,60.0875015299796,60.0651016199815,60.0708351099789,60.0145835899663,60.006248469954,59.9929199199553,59.9762191799474,59.9884223899219,59.9854736299129,59.9726333599114,59.9987220798965,59.9912719698867,59.9755210898886,59.9593200698667,59.9712982198496,59.9575462298172,59.9666747997591,59.920532229754,59.9221763597768,59.9088821397734,59.891979219737,59.8787231397369,59.8688964796832,59.8383178697053,59.8072738596942,59.7877769497097,59.7691650396849,59.7586097696991,59.7391662596867,59.7294425996949,59.7414359996994,59.6913871797352,59.6964721697027,59.6832771297039,59.6782836896614,59.6402778596583,59.592498779686,59.5860099797026,59.5990447997021,59.5862388597226,59.5161132797619,59.4869079597652,59.4838905297402,59.4627761797485,59.4627838097282,59.4210815397183,59.392272949732,59.3911094697123,59.3610343897142,59.3488883997163,59.3463897697557,59.3121490497734,59.3082885697433,59.3195190397302,59.293884279711,59.3250122096949,59.3274955696712,59.365295409617,59.3555297896206,59.341388699569,59.3058547995722,59.3157768195531,59.3069457995437,59.3375244095307,59.3344840994991,59.3024215694923,59.3063888495166,59.2838745095147,59.2288894694841,59.2395019494971,59.222286219498,59.2327766395285,59.203334809527,59.2430572495716,59.1898345895813,59.1973266596053,59.2199897796026,59.2269439696201,59.2061157196484,59.1666679396643,59.1100006096499,59.1400756796279,59.1047477696181,59.1247215295804,59.0952758795606,59.0647239695971,59.0211753796148,59.076389309645,58.9029197696144,58.876110079635,58.9027786296577,58.8096923796699,58.7958335897063,58.811279299718,58.7891654997195,58.8088874797325,58.8790893597393,58.8936767597335,58.8831176797261,58.9458503697239,58.8962593097365,58.9915237397413,59.010833739751,58.9609298697516,59.0294799797694,58.9588928197681,59.0567016597956,59.0498771698029,58.9772224397975,58.9828262298028,58.9574584998113,58.9564056397829,58.831115719774,58.8280563397882,58.8833351098094,58.8255538898055,58.8072204598341,58.8322753898482,58.7677764898581,58.7327766398449,58.7833824198141,58.6875266998166,58.7083129897926,58.7371368397897,58.7063903797838,58.7100830097713,58.6874999997733,58.6773071297903,58.6440963698001,58.5819435097813,58.7163886997303,58.6650810197143,58.7674140897018,58.7874984696644,58.7736816396636,58.7857055696539,58.7700004596469,58.799720759633,58.7830810496288,58.820835109616,58.8188896195954,58.8504219096,58.8731079095838,58.8767089795256,58.903930659497,58.8994750994781,58.8831253094794,58.9020156894596,58.8730544994633,58.9044799794139,58.8930702193404,58.9052734393303,58.8953247093244,58.9069442692774,58.870555879298,58.8825073192665,58.8808326691468,58.9199790990329,58.8925514190075,58.9108886688641,58.896118158862,58.8983344987915,58.8591651887224,58.8696899386997,58.8191680886678,58.8356018086177,58.8161010686175,58.8172225984863,58.7894439684986,58.7811279284042,58.7222213684607,58.687861793859,58.6983337383425,58.6889038083261,58.6873336798041,58.6800003083531,58.5961303683779,58.5717163082406,58.5283317582838,58.535556788169,58.5570144681089,58.5508346580641,58.3842391981106,58.243476868297,58.1847228983035,58.1686172483559,58.124118798284,58.1955642682752,58.2518081681653,58.2934608481819,58.3665161080522,58.4414672880116,58.4838867180546,58.5216674779778,58.5319213879251,58.50656127779,58.5214004477455,58.5102767876826,58.5839576676452,58.5516662575104,58.5433349575655,58.5372238174959,58.5183334375446,58.5105552674533,58.5230712874208,58.5024986273577,58.451110837395,58.4311103774904,58.4347228974243,58.397666927434,58.3845214773618,58.3752784674073,58.3380737273108,58.3466796872591,58.3353271472162,58.3091659471845,58.3153076172645,58.2933349572191,58.2438888473251,58.2344436575137,58.1278457578426,58.0758743278401,58.0928573578201,58.1243286075934,58.2244872974425,58.236946107224,58.2269172671905,58.2574996871997,58.2955284070582,58.3088874771366,58.3905029271753,58.4296874971375,58.4344177170317,58.4716682370625,58.465892786897,58.4516677868204,58.4391670168747,58.4513893067894,58.432777396793,58.4224853466752,58.4105567867817,58.3922882067685,58.4050903268211,58.3596801768201,58.3450012167418,58.310832976889,58.286071776756,58.2683334368202,58.2822875966814,58.2641792265187,58.1408348067363,58.1041679367028,58.1005554166193,58.224723816302,58.2302780161524,58.297222135814,58.2913894655595,58.272468565667,58.2719116154748,58.3230552652605,58.3438873249938,58.3275146448609,58.3386573747519,58.3711509645496,58.3991661045653,58.346389764106,58.3864173838694,58.4199981640468,58.4203187489053,58.4341201739307,58.4300003038186,58.4203603910903,58.4208335836782,58.4960937434029,58.4961128132033,58.5122756932688,58.47472381283,58.4525146430097,58.4202766327004,58.4550018227157,58.4627761725771,58.4483337324428,58.4683608923257,58.4963874724251,58.4947204521296,58.5111122019191,58.4827766315804,58.4913902212822,58.4741668610901,58.5466651814165,58.5567016512643,58.5463905210692,58.621948231087,58.6163902209124,58.6327781607156,58.6723823408306,58.664489740643,58.7022209104441,58.7160224806084,58.7094459403575,58.7265395999433,58.7121543799817,58.7133178597645,58.7427787696995,58.7396850498725,58.7605552600445,58.7763900698097,58.819126119931,58.8301429598614,58.8197021396419,58.8363876196149,58.8264770392946,58.8530654792538,58.8402709888508,58.8116683889704,58.8196449184833,58.7941703678579,58.7277641171443,58.7455558671113,58.7276420466842,58.6492919760972,58.6425018155474,58.6613883857102,58.6694946154175,58.6544456351288,58.637882215365,58.6305541851356,58.6319427353934,58.6230544845484,58.5405540348422,58.5263290253662,58.4975128054763,58.4960937347555,58.5147094548229,58.5183181642971,58.5302772343823,58.5217285040263,58.5966682241132,58.5769462432517,58.6291656340172,58.6480560146911,58.6917114147199,58.6838874651688,58.6975097559697,58.7164916858826,58.7141685356624,58.736511216088,58.7276382256458,58.7627372558149,58.7791671661295,58.7545204061505,58.7639160064709,58.7938880765782,58.7944450263326,58.834106436257,58.8375243958513,58.8616676158995,58.8655547954616,58.8255882150264,58.8444175547348,58.8271293440938,58.8403663439396,58.7879447734134,58.8551025234068,58.8574828931514,58.8341369429811,58.8576774428417,58.8866958444959,58.8718757446211,58.9127769355593,58.9550018135871,59.01672361406,58.9880981236812,58.9835204928463,58.9352760113988,58.9738998214328,58.9963874627016,59.0180549430529,59.0312652415729,59.0338744929076,59.0458984229012,59.0419425822868,59.0604858218022,59.0472221209978,59.0149993706756,59.0097236402032,59.0208740001,58.9958343291797,58.9283904775798,58.9217338369662,59.0119437991263,59.0045089494064,59.0297240997246,59.0263900603724,59.0405273198681,59.0394897303505,59.058715800517,59.0587043597604,59.0714721497662,59.077144599081,59.0394439483814,59.059326148557,59.0410766369294,59.0583419569962,59.0441665362034,59.0672225767192,59.0649986077881,59.0863876092078,59.1175079098857,59.07975766956,59.0702781510976,59.0947532514085,59.1005554011332,59.1838874627354,59.1889037924751,59.1727790622637,59.1983451622448,59.2042846512818,59.212925008179,59.2067260525652,59.2172851428411,59.2202758625959,59.2138515719141,59.224979382344,59.2380561628126,59.2383346422422,59.2703246927484,59.2591667020603,59.2849998324171,59.2736129617771,59.2530555517112,59.2647209015561,59.2544555511735,59.180000290685,59.1483344800993,59.2388877707891,59.2611160108511,59.2597045705262,59.1455573793105,59.1402778388212,59.2536125001698,59.2830543300973,59.2494430291465,59.2669448690208,59.2705573796418,59.2908324994802,59.2983321901956,59.3430862204397,59.3414153898969,59.4026565414125,59.4030647107797,59.3829002206928,59.3861083801808,59.3524780094715,59.3805541799505,59.3688964593706,59.2427787564159,59.27694318684,59.2819709560889,59.2833327971196,59.3394431878114,59.3491668487068,59.3833007585296,59.3777770781596,59.4097289783887,59.3858337169898,59.4163894477917,59.4302787594288,59.4520492396491,59.4386634586567,59.4631080391011,59.4641685297373,59.4838065897333,59.4780883586556,59.4548949980929,59.4580650077767,59.4755706583354,59.4774894480088,59.4013900556697,59.491920447927,59.4786109664426,59.4935302462815,59.4753722858936,59.4650001241584,59.442501044395,59.3977775330895,59.3741683701857,59.4094848329413,59.4422225739804,59.4658813238785,59.4786109643357,59.5081176563892,59.5005340378908,59.5592155286743,59.5849990586127,59.6674270398916,59.6583251795422,59.6699981494219,59.6997070101707,59.7463874602787,59.743103009787,59.7672157096195,59.7527770791517,59.7913970695641,59.7924995187099,59.8102760087834,59.8161430183972,59.7888412278318,59.8025016580574,59.8086127976021,59.8236236382544,59.8213882178708,59.8411254676493,59.8311118874764,59.8638915771772,59.8218993861356,59.8363876061748,59.8236121853577,59.7747192144474,59.792335483962,59.7772178334403,59.7869262433432,59.7977790544388,59.8166656246296,59.8097228748299,59.8349990556183,59.8419182322885,59.8408813257434,59.8386115764733,59.8574981469701,59.8685455067697,59.8552332391127,59.8847121964453,59.8603706152009,59.8864745858367,59.8919524851538,59.8620834145106,59.8793220244262,59.8665275339522,59.9094848346526,59.91638944498,59.9077758547455,59.9030761451343,59.9213180257929,59.9347305055079,59.9135474945155,59.9289207248622,59.942222574299,59.9660186542906,59.8975143117247,59.9724998240066,59.9925193539678,59.9738136225466,59.994110083169,59.9836120333095,60.0026015942395,60.0499992142652,60.0624999740279,60.0505218236246,60.0624999736657,60.0437507330564,60.0598983532507,60.0562515027736,60.073440522315,60.0875015032183,60.1249999725935,60.0901069311452,60.1557311721311,60.1604156216114,60.1463508313717,60.1869811714425,60.2083320306206,60.2026061700265,60.222393010193,60.2130241093133,60.2291679100121,60.2171897611319,60.2562484413472,60.2791671506743,60.2630195301476,60.3098983493861,60.3192710584752,60.3276061691257,60.3703155193486,60.3541679087217,60.366664858533,60.3546724201211,60.3359374671295,60.3416671467187,60.3041648543093,60.3328132250888,60.3354148561877,60.3619804070401,60.3437499647812,60.3009681324976,60.3032951065111,60.258468599564,60.2281951595554,60.1881560972177,60.1686286571001,60.1811561258001,60.1670150440493,60.1170310637754,60.1054153046207,60.1083526274589,60.0846099561927,60.0561294268767,60.0614661904832,59.9703178087386,59.9744872668497,59.9547309568279,59.9588737156901,59.952556575017,59.9301299651383,59.9302940042095,59.8772010453684,59.8860320669117,59.8696746471041,59.8555678964667,59.8528480173187,59.8256072682427,59.8032722174639,59.7951507279741,59.7603034674361,59.7296943378089,59.7172393465333,59.6759986580369,59.6621551193118,59.5915374486311,59.5931777699606,59.5670470901684,59.5469245587623,59.5533408781751,59.4469070046567,59.4819717035291,59.4661826726654,59.5012969607572,59.5213737107371,59.5419730691051,59.5127753763146,59.4974593663837,59.5029525369479,59.4737967978345,59.4591750695566,59.4229812207814,59.4121779995447,59.3956298411322,59.3707809013303,59.3495750002289,59.3104362113594,59.2571105599695,59.1967429706724,59.1803931794635,59.1064185699627,59.087604477938,59.0926055469121,59.0672988464816,59.0656318252953,59.0129546637893,59.0004996846357,59.006183577067,58.9815177488475,59.0194320298228,59.0025596204,59.0350455821712,59.0386123338756,59.0746955542149,59.0749587663056,59.0324859275454,59.0203017872947,59.0154075280241,58.9564094172895,58.938384978425,58.9370002373985,58.9168929769861,58.9238013858245,58.9507827450972,58.9437522533259,58.9075164424309,58.8874549508862,58.9082183386226,58.8902778177142,58.9038199966705,58.8918761759171,58.9045638554762,58.8967513542768,58.8641166227648,58.7836493925149,58.7890395611141,58.746166179878,58.7559966484393,58.8103484573218,58.8137092058434,58.8302535452769,58.8319777817837,58.8470611002268,58.8840598492023,58.8771743174785,58.8870505669909,58.8547095652759,58.8571776630381,58.8365516014356,58.7620467382742,58.714641503861,58.7252234749395,58.7063865068337,58.7167357780504,58.7032851593231,58.7101172806704,58.6964415912611,58.7102546130163,58.6852454542094,58.702629037166,58.6396521988226,58.5632285585949,58.5234488864293,58.5296363246096,58.5588073138413,58.5716552120077,58.5465812107648,58.5045127309622,58.4897460292891,58.4294394815198,58.444869942912,58.4443587748395,58.3659820060243,58.3710326597976,58.2874526513163,58.247734021273,58.2274207620713,58.2083816054582,58.1866721666856,58.098373366914,58.0611571862619,58.0693015647314,58.0445174732152,58.0614127624141,58.0387191318693,58.0369834409532,58.0250319915032,57.9895438611311,57.7700232962221,57.7644042448037,57.8095206647249,57.7885512721048,57.801120701287,57.7126082801851,57.7190245990164,57.684505397508,57.7207450258639,57.6742781955969,57.6526946336144,57.723941732901,57.741245200399,57.6638373602995,57.6419677011244,57.6034354532322,57.6153449337791,57.6011313748327,57.5794867853064,57.5924491262349,57.5357436454134,57.4935721761574,57.4393309855748,57.4265631945189,57.4382094640176,57.3683318433496,57.353198946245,57.3322867667607,57.3343009285883,57.2776183494054,57.226123748772,57.19625847929,57.2041892380307,57.2621573768859,57.2517279951716,57.221595694328,57.1960181554578,57.1399268451452,57.0788802466543,57.0860823993049,57.0127753592464,56.9992141080801,56.9776305599255,56.9041594899403,56.8968238181186,56.8687400179133,56.8773040192812,56.8617438726546,56.7786788328306,56.7670020549038,56.7464942347936,56.6997565668591,56.6538314251214,56.6530074530391,56.6063498937894,56.5337600098134,56.4662055397025,56.4690627481308,56.4419593177691,56.4082106916765,56.4411124667574,56.4252433275052,56.4021033655583,56.3699263959052,56.3336333649381,56.3034057064647,56.2798995446302,56.256889284504,56.2678145824369,56.2563971906247,56.2711180995746,56.2635344888268,56.2308310887344,56.2125891970559,56.1548804627406,56.1027297415782,56.0701445925952,56.0911483197798,56.1371383067187,56.122497496078,56.0928611177371,56.1006240160952,56.0846328050745,56.0450438859584,56.0716857216153,56.02352516866,56.0400122832846,56.0295142312019,56.0098838030572,55.9877776322615,56.0058860058384,55.9674758181178,55.9721907892523,55.9481772704556,55.9535407313548,55.9305152217443,55.9314612642118,55.9100112263555,55.9044493953241,55.8790740359903,55.8465003326763,55.8251380225415,55.8130263636183,55.7899970331586,55.7877883194284,55.7593268596742,55.7382163315481,55.7315635005249,55.726638721507,55.6786002407015,55.6715087223828,55.6515883728429,55.6576728110802,55.6415785983703,55.6254805799957,55.5988883290247,55.5087126999823,55.4672393136253,55.4230498588991,55.4292067716645,55.4003829198654,55.4183615764771,55.4118041151286,55.3660468252714,55.344058910083,55.3396033453184,55.3207205959296,55.3254965936691,55.3098677832491,55.2962569468572,55.2668875892353,55.2519110882088,55.2511557762431,55.2426070470488,55.2301444265193,55.243404314763,55.225467604118,55.2552642007844,55.2490080983413,55.2019461803077,55.1876143646775,55.1681441531191,55.1557501950472,55.1402625247052,55.1287345161347,55.1035498854509,55.0689467665943,55.0344428242354,55.0083922561421,54.999874034242,54.978794024827,54.9770430762948,54.948863906276,54.9299544549556,54.9199370562809,54.896591115447,54.909202506978,54.9045676490311,54.9253386794871,54.9558944030298,54.9420546843863,54.9261359532844,54.9145125646419,54.8760566051357,54.8169211638131,54.8010787368592,54.7888030364008,54.778407987545,54.7256430932161,54.6736831017149,54.659797600249,54.6249503397973,54.6507834735253,54.6007041264859,54.6201171291417,54.6021232001269,54.6009063117984,54.6229552628565,54.6131896435362,54.6555861834201,54.6935882059226,54.7213591958472,54.7310256492333,54.7533606994421,54.7904929624593,54.7910613551162,54.7521514445998,54.7617911862184,54.7448691867589,54.733524276108,54.713661146796,54.7357139199836,54.7235908104613,54.7359695013429,54.7300948721824,54.7557754129427,54.7673225017829,54.7881469323691,54.8108443817182,54.8295287720269,54.8430785726425,54.8325690829732,54.8490485733385,54.836334193665,54.8410377143932,54.8762702541099,54.9200057656437,54.9373969669037,54.9326247878846,54.9695624993201,54.9614791601401,54.9281921102876,54.9308509511929,54.8479575817284,54.8443565131068,54.8175697033493,54.8519668340776,54.8187598944249,54.8299445849361,54.8198318253328,54.8418388154063,54.8492431361754,54.8343772666877,54.7841834757866,54.7481155156812,54.7285842661241,54.7302741777959,54.7531852491949,54.7081641998739,54.7867927408183,54.8507995421272,54.9272117421349,54.9424819744444,54.9190864445546,54.9855155758581,54.9730071860266,54.9918060167597,54.9754714868714,55.0148353471032,55.1409454188506,55.2033767592954,55.1953239295032,55.2353820697624,55.2781524603755,55.2551460206993,55.3120384110332,55.3567428518377,55.3232765117764,55.3249473519744,55.3069686819443,55.2271232510751,55.1774864107813,55.214992511436,55.1531791613575,55.1441764715812,55.11841582167,55.1135711614213,55.0695152209589,55.0906028614087,55.0440521112635,55.0255470207717,54.9838714503963,54.9746246205934,55.0046462910585,55.0000152514896,54.9236640808354,54.8979644708969,54.8135032601852,54.7702178906427,54.8132018912541,54.7667388811969,54.7449150007992,54.7096710107128,54.8160095122512,54.8021736025959,54.8619461032752,54.8949394134203,55.0242118746276,55.0455169645808,55.0512275644294,55.0764274546317,55.0886344852919,55.0628585755318,55.0085029554498,54.9889564452501,54.9578590352381,54.9396858150399,54.8338127045442,54.8249130143459,54.8009262043483,54.7639579741044,54.6929321234418,54.6737060434917,54.6577186538254,54.6245384138772,54.6095352140841,54.6151924043745,54.58441924439,54.5917930546364,54.5651740945743,54.555877684747,54.5511436445414,54.5382575945614,54.519557944354,54.4866294845904,54.5087203946791,54.5051002449001,54.5181503248779,54.5174064550143,54.5692329353488,54.5849113456264,54.5709495457747,54.5274124055471,54.4815063456584,54.4941101059952,54.4657669061041,54.5093879662521,54.4538421563264,54.4438743564324,54.4574012765437,54.4368133466806,54.3868560765311,54.350261686612,54.3119239763661,54.2469901963669,54.1957549961173,54.1885757362621,54.1608047462271,54.1570014966645,54.1281356768165,54.0996627767799,54.0915794366385,54.085521696742,54.0412788367497,54.0200118968477,53.992309566698,54.0000038066233,53.9797401365493,53.9719085663593,53.9302825863134,53.9216957060316,53.8692474360806,53.8586044259004,53.8411445559194,53.8366889956206,53.8012084957818,53.7954635560511,53.7802925058693,53.7626991260544,53.7572708056275,53.7287445056949,53.7088813754735,53.7065010053326,53.7241935653644,53.7251357952603,53.6146545351495,53.5972099250648,53.5841522147552,53.5529632547796,53.543521874616,53.5573883044673,53.5303382842615,53.5327529840999,53.5530357340506,53.5302085837664,53.5287589932058,53.4961471528878,53.4642372029514,53.4731368925753,53.4186592024829,53.4130592222493,53.3952941823621,53.3909301726274,53.364620202756,53.3679656930104,53.3526191629541,53.3346443131429,53.320079793091,53.3064536927036,53.2904128926699,53.2132644628279,53.1816940227069,53.1600265430523,53.1384506129354,53.1371078426994,53.1174621529073,53.0843582128976,53.078872673389,53.1395492532716,53.1393127334315,53.105113973673,53.1378593337796,53.1496658242439,53.1812896644848,53.1589240946881,53.1402854846135,53.1177368148867,53.1320915149048,53.1218833849948,53.1320266649801,53.1319770750923,52.9961891149847,52.9811973548967,52.9795188843833,52.9638137745467,52.9654083247784,52.936664574928,52.8778266847923,52.8777084345073,52.8417701642899,52.8174591039245,52.8544425938592,52.8754539433518,52.8372383032565,52.7777366533843,52.7470169031723,52.7595977726299,52.676673882259,52.6747856019546,52.7274589422814,52.7173080319893,52.7683753920199,52.6662483113237,52.7515029812427,52.7974319414919,52.7635765009784,52.7637138307181,52.7910613906699,52.783657060402,52.830905900547,52.8726425103879,52.8903236305367,52.8975601102844,52.921890250432,52.9542579602294,52.9370002597583,52.9639549193211,52.9463691591476,52.9974555890764,53.0001296887938,53.0425338589164,53.0551872187712,53.0211868181458,53.0247726279185,52.8691139067613,52.8601341071469,52.8333930870274,52.8774833580022,52.8583373883584,52.7944564680192,52.7954177776507,52.7414893975237,52.7286071672837,52.6846313375921,52.6372070172331,52.6344642469599,52.6174125569859,52.6000594970962,52.6045913572871,52.6822852980832,52.6364021185769,52.5520438979082,52.5308990378781,52.5287742481826,52.5097122078609,52.473220817753,52.48636625813,52.4761123581225,52.4307594177847,52.413040147924,52.3972091576133,52.3604163976363,52.3513755678339,52.3830222983795,52.3501319786511,52.3416366490386,52.2948760886896,52.2846679588387,52.2661743085028,52.2556571888539,52.2185363684924,52.2298393082374,52.1766013977641,52.1528167579574,52.1507072275271,52.1318168474293,52.1503677268526,52.2855186373166,52.2853736770154,52.3091201670534,52.3100776568393,52.2269897361283,52.2134666258116,52.2305030655836,52.1985015750277,52.1550292846973,52.0865707248959,52.0977706745829,52.0799255239659,52.0547370737244,52.0565032833989,52.104637133282,52.0760688624536,52.110408762176,52.1185607711908,52.1056976110787,52.1078300304452,52.0828704601673,52.1172752191863,52.0809211488855,52.0552596789271,52.0415076993642,52.0076827793267,51.9868087589123,52.0430106884517,52.0328063777997,52.0445594571613,52.1104545364885,52.0502052064352,52.0110854857596,52.0021095051891,51.9655532547782,51.9673423542693,51.9958762842685,51.9811019640502,51.9424171138748,51.9018935947762,51.8673324344915,51.8642081941184,51.8227538840693,51.8290252435544,51.8700332334014,51.8738441226601,51.8614730520977,51.8223838521958,51.8127937011457,51.8220786704515,51.7656211606708,51.7459716488554,51.7696494779707,51.7651404971289,51.7585715873565,51.7154006658303,51.7366714149013,51.7644309655511,51.7778586952348,51.7625007234615,51.7781409824514,51.7531356416723,51.7559547001031,51.7075538196504,51.7050323083228,51.6289863179227,51.6328773073143,51.5738982759163,51.6246222933316,51.6761016324045,51.6628913414836,51.6652945998982,51.742294257985,51.7569732188295,51.803642218841,51.849277450338,51.96765513004,51.9790801505068,51.9891356899402,52.005493110572,52.0048636894379,51.974750468147,51.9779586268232,52.0776481084995,52.0797461972648,52.1447219272128,52.1160582945854,52.1460112944066,52.1770133456749,52.2248801660778,52.2240333067179,52.257102916205,52.2753943871082,52.2845458459624,52.3130301861641,52.289699495387,52.3066558246837,52.332721655109,52.338954874329,52.3630637545523,52.3940581727382,52.4142531727441,52.4245337835787,52.5199241041339,52.5819854164772,52.5772056058534,52.5957717355042,52.584251344768,52.6034354642258,52.670562684824,52.6864242033672,52.7267074037888,52.7365874617957,52.7232360209273,52.7382430403839,52.7310256369515,52.7577895463268,52.7500762252301,52.7739638646725,52.7585295927122,52.7733344309874,52.7703093790779,52.8015326789347,52.8237609161124,52.8520545273776,52.8441580947769,52.8774069951766,52.8395499438682,52.7741851050189,52.7275847614668,52.7135542982572,52.659362705456,52.6455191773804,52.634704520754,52.6056823070137,52.5683288002033,52.4660720222263,52.400978029785,52.3625945395133,52.3224982546307,52.3280524508301,52.31308738983,52.3140296233343,52.2838591808725,52.2753524120093,52.2606543801569,52.1654586082067,52.0876158982797,52.0830992973377,52.0424040961323,52.084731992846,52.064998563437,51.997848442269,51.9958112946818,51.9965131413574,51.9969823965683,51.9984748204801,51.4251105577761,51.4244467730684,51.4063879672224,51.4119383182347,51.4592557956108,51.5048833878837,51.4892431266207,51.4993101106631,51.4772228874852,51.4616322098884,51.4832043056687,51.4663585869156,51.4680828533441,51.4422420945877,51.4348758898162,51.4584583550865,51.4652332371523,51.4337734183818,51.4472125927619,51.4322665880298,51.4394267584567,51.416759801597,51.4225695738611,51.4675029171591,51.4291499360962,51.4190485718474,51.4353335080173,51.4436457495483,51.439090986238,51.4584963453429,51.493809019914,51.5175173372517,51.4813502116849,51.455074556353,51.4574625408254,51.4717981788926,51.4651987462771,51.4460337113221,51.4189989273512,51.41818258658,51.3955995863843,51.4086114947357,51.4363519889583,51.3094216430633,51.3045693290485,51.3262635234717,51.3007126691229,51.3634262959748,51.2946549250157,51.3045235428471,51.2788887453005,51.381504125565,51.2796211591812,51.2664489911494,51.3165779316169,51.2666664296525,51.2650299229,51.2917404244878,51.3267517197105,51.3358344962128,51.3602790773103,51.3697204491715,51.382686613737,51.3924484318119,51.3689002796913,51.3455085614209,51.3520164409382,51.322734824766,51.3168487511893,51.3272285517637,51.3103179958562,51.319778432151,51.2929496803908,51.306446071871,51.263946525758,51.2753829745217,51.2913055096481,51.2771110161832,51.2978095599369,51.2845344065887,51.3139075714184,51.309684687964,51.2347144214813,51.262462530828,51.2314833692561,51.2766684535064,51.2729720083607,51.234676263173,51.2228812181687,51.2338255724099,51.2630537821787,51.258586767906,51.2228583219619,51.197776694258,51.1972235591194,51.1789054055034,51.1678923705891,51.1775130242508,51.148456472614,51.1271590126219,51.1388396114243,51.0911101065277,51.0795820776251,51.0997236847746,51.0720442350064,51.0606611812361,51.0876959375854,51.0541647475822,51.0380552721115,51.053611619368,51.0258329904258,51.0169447424391,50.9795645188834,50.978870234778,50.9774664306966,50.9555242892507,50.9599989263661,50.945278018401,50.9561117564654,50.9449652041272,50.9812582335841,51.0313871799679,51.0587690772425,51.0402792364646,51.0552061435578,51.022861318376,51.0347936866868,51.0105627361087,51.0141676211392,50.9991300845936,50.9780691400758,50.9955557056709,50.980277844964,50.9814603988212,50.9667318566943,50.9693983306789,51.0008314366656,50.988178089778,50.9455564743283,50.9412191685044,50.9255636486003,50.9272230460052,50.9098089486032,50.8822210535122,50.886176899453,50.869270159622,50.8761099153099,50.8549001945735,50.8359182666566,50.8375013698586,50.8155554096326,50.8041647243603,50.7684286302518,50.7861173772012,50.7745664742425,50.7734831076309,50.7386091411265,50.7527197024649,50.7719534035693,50.7697637704063,50.8170049790947,50.7831719009002,50.7936094389629,50.7684820299028,50.7536466673681,50.779723938026,50.7778744319176,50.7675550544593,50.7581327363053,50.7426794092833,50.7224996571761,50.7405469878538,50.7255132655779,50.7561719846816,50.7577779718057,50.743331711635,50.7566678908627,50.7095640123475,50.6983335362071,50.6486127747276,50.6581952882815,50.6257055097188,50.6100003978554,50.619563846196,50.5622213154435,50.5692517973747,50.5461118492233,50.5332906455606,50.5428388337155,50.5602758141048,50.5576513031456,50.5175016124399,50.5220449207539,50.4724576696579,50.4660794886858,50.4888875688904,50.4645078379586,50.4666669562603,50.4435460744671,50.4511487636681,50.4325673736123,50.4236104623244,50.427848591806,50.4512746420484,50.4074552113815,50.3884236905712,50.3807866708802,50.3711125901677,50.3577764105167,50.3639104515399,50.3522222106102,50.3422238911139,50.325000530859,50.3291661815773,50.3381650517049,50.3311116812255,50.3525426516049,50.3360593420788,50.2811887303473,50.2988889303268,50.2745320901665,50.2730519898898,50.2527768699942,50.2683980497033,50.2820661198845,50.2900006897351,50.2796590396324,50.2912671695911,50.2497785196994,50.2444455694778,50.2774045595107,50.2834470394167,50.2544438893793,50.2427785494746,50.2252767194938,50.2294423694253,50.2400014494077,50.248611219363,50.2419431293961,50.2209889994835,50.2488896893788,50.1936109198623,50.2280538196689,50.2099988500379,50.2252767199469,50.2688901494486,50.2411115298194,50.2552793098557,50.2836110694557,50.2680547297988,50.282256849692,50.28277564983,50.2247235906122,50.2216680112398,50.2716672503804,50.2458455609819,50.3025014509359,50.3277776302008,50.3441655707802,50.320834881325,50.3247220608922,50.2855565613783,50.2827909110509,50.2636106114386,50.2530553411987,50.2227780915803,50.2417867317457,50.2087667121087,50.2324979419101,50.2363889327197,50.2094457228292,50.2477758036214,50.2269437442543,50.2463872542752,50.2374990156573,50.2547223758184,50.2082975071566,50.2108342785276,50.1999204287697,50.2099988597408,50.2402951898312,50.2186124500694,50.210918211265,50.217498561873,50.2319448213362,50.2134549820271,50.2248418524959,50.2147214624845,50.2148778635669,50.2316663431119,50.2377774939525,50.1933629762901,50.2191655873909,50.2020642987324,50.2413900173069,50.2566678750805,50.2605550555954,50.245834138029,50.2427785674857,50.2060048896354,50.2213895599259,50.2088888006786,50.2181508810751,50.1832998120831,50.2141683426446,50.2222211616625,50.2374990321377,50.2419431508313,50.2311094144572,50.2602765857136,50.228053844389,50.2358320025623,50.2208326124536,50.2008321632276,50.2266233362985,50.2109449269693,50.2548330184023,50.2793729707493,50.2717893523937,50.2745664504511,50.2529828884891,50.2323415691079,50.211883337555,50.1760557075106,50.2067449495889,50.1896818103789,50.1858671093716,50.181041520252,50.2040899229316,50.1743238432226,50.18956355378,50.1887624655224,50.1981504348103,50.194812577356,50.1743238475453,50.1923368487897,50.1895940805296,50.0855901926164,50.1102064370691,50.105884384214,50.1331747305258,50.1390798771407,50.1578977743705,50.16213331293,50.1535604654003,50.1816156931682,50.1456754873626,50.1281507759901,50.1273420569255,50.1818998575726,50.1937330564728,50.1879995684686,50.2032964975649,50.1955145197726,50.212116078389,50.2232588102335,50.2083433511078,50.2270506216818,50.2141607633481,50.2371137928362,50.2214468335694,50.2309721352588,50.208331907313,50.224445188757,50.2161100800739,50.2360571358533,50.2294424588934,50.2638891729687,50.2555578743855,50.2794111858352,50.2627791070571,50.276389948194,50.2586096389,50.2911108594151,50.2520559912403,50.2736128536249,50.2575796752949,50.2816656767569,50.2641600377682,50.2708319414073,50.2588881201141,50.2736548221968,50.2671583955045,50.2861098064174,50.2749213078352,50.3113898046003,50.2885664779235,50.3097647480187,50.2847212589084,50.2750357510474,50.2750014232227,50.2891653932338,50.2727011574661,50.3149794498825,50.2983321117922,50.2913893706364,50.27638998166,50.2734602942767,50.2883681364063,50.2772215879323,50.24719228629,50.231624508449,50.2422217390287,50.2205542592711,50.235053935236,50.2008322870167,50.2033690589885,50.2433699796942,50.2524146207736,50.256423873069,50.2363890853641,50.2657889568061,50.2516669484406,50.2883338194874,50.2860106726123,50.3077773322432,50.2894439045524,50.3019027073671,50.294444979133,50.3069075895224,50.2953332657006,50.2950307071803,50.3058318506652,50.2948358918075,50.2908324626409,50.2552031950619,50.2829436804215,50.3193282637895,50.2836112561821,50.2794456101508,50.2661094313123,50.279445612278,50.2602767544274,50.2716598156217,50.2737871811933,50.263053856304,50.292499509766,50.2874946318707,50.3046073620572,50.2942504635036,50.3037605041081,50.2546996896982,50.2569427304213,50.2714767305415,50.2552795206933,50.2589912212547,50.270675641319,50.2502403115681,50.2905540332934,50.2619132840068,50.2030563246235,50.1861114352714,50.2133331166539,50.1858329679534,50.2210807684007,50.2296180582415,50.2627792283798,50.2694434988224,50.2112502995875,50.1602783093674,50.1624984585387,50.1272010688028,50.1502761697809,50.1105537309708,50.040378561201,50.0297241117486,50.0172233517898,50.0202789223301,49.9814376727598,49.9213333029288,49.9180564828057,49.822776783319,49.8183326634084,49.8408317534251,49.8474998435929,49.8083343439342,49.7994461038737,49.7699667125724,49.6686096140404,49.5941657944173,49.4844436543784,49.4219512846599,49.4183998047786,49.3916664047889,49.3161125151578,49.3252792360555,49.3083343461098,49.3161125162049,49.3088874761217,49.3136100762704,49.2997207563726,49.3147239667859,49.3033332767097,49.2683334368804,49.2919425971158,49.2794456475038,49.2949981675629,49.2533645576381,49.2707748377622,49.2019462578069,49.1791915880844,49.1903228782099,49.156627658011,49.1591148379132,49.0994453379198,49.0927772481873,49.0458335883598,49.0441284184836,49.0961112983495,49.1363601683167,49.0799789384189,49.0735168485881,49.0528945886338,49.0625953686484,49.0502777087112,49.0159568787539,49.0082244887347,49.0041656487681,48.9330558787757,48.9350013688517,48.9113883987989,48.9225578287422,48.8997230487713,48.8922233589765,48.8387756290592,48.8394432090836,48.8198318490907,48.8228645291625,48.816112519146,48.8119430491757,48.8061103791514,48.7677764892064,48.7869453392163,48.760749819238,48.776943209229,48.7687225292891,48.7439651492711,48.7005805993096,48.5953903193214,48.6325492893199,48.5628433193672,48.5815238994256,48.5737190194361,48.5132713294766,48.4705543494782,48.4140663095066,48.3528938295625,48.3367233295547,48.2996711695764,48.2655143696202,48.2405319196303,48.2284469596506,48.2420349096496,48.2155494696602,48.2363357496637,48.1843223596809,48.1374778697134,48.1399040197292,48.1089286797342,48.0977134697252,48.0672721897462,47.9738883997548,47.9591674797695,47.9480552697623,47.9146957397651,47.8594436597888,47.7698516798022,47.7538986198188,47.7031173698292,47.650222779865,47.661079409872,47.6222915598683,47.5793876598818,47.5235557598827,47.4879989598887,47.426055909927,47.4417343099328,47.3846664399333,47.3310546899401,47.2738342299428,47.0871658299603,47.0499420199654,47.0299568199723,46.9719428999785,46.9280548099808,46.8855552699844,46.859722139985,46.8488883999864,46.8041648899869,46.7505569499893,46.7217636099941,46.6694717399962,46.6545715299971,46.6768112199983,46.599891659999,46.5505943299995,46.5003280599996,46.4489364599997,46.3823776199999,46.3132438699999,46.27046585,46.27456284,46.24355698,46.18946838,46.17159653,46.10121155,45.98117447,45.91001129,45.71466446,45.70274734,45.64559937,45.55593109,45.51403427,45.51036453,45.54820633,45.6189537,45.69611359,45.64136124,45.53586197,45.46123505,45.42151642,45.40982056,45.43621445,45.4135704,45.40246964,45.4158783,45.4682579,45.46729279,45.48407364,45.51252747,45.50965881,45.49036026,45.49071884,45.52184296,45.53188324,45.50997162,45.42881012,45.39276505,45.3733139,45.3483696,45.30632782,45.30344009,45.25666046,45.25573349,45.19869232,45.3026657099999,45.5661087,45.5728340099999,45.5937309299999,45.5948333699999,45.6385497999998,45.6280631999997,45.6449966399992,45.5938072199988,45.5767478899984,45.5868797299976,45.5369834899966,45.4608497599907,45.4246635399901,45.4123916599886,45.3726997399866,45.4026374799836,45.4796447799795,45.5138511699738,45.5188789399663,45.5078277599639,45.468608859963,45.4578323399521,45.5190391499367,45.5318260199219,45.5606346099136,45.6246643099116,45.6654815699045,45.6809425399084,45.7091598499077,45.7346038798938,45.785282139891,45.8509864798941,45.8727073698908,45.898147579866,45.8922576898581,45.8540420498644,45.8468627898583,45.8036613498619,45.7890853898566,45.7866363498431,45.8355827298157,45.8638610797794,45.9488906897453,46.0155143697445,46.1592216496025,46.1987647995334,46.1826972995209,46.1975707994475,46.2489471392696,46.2438468892143,46.2745056191337,46.2739448489775,46.2523689289099,46.2916870088018,46.2945899986249,46.318851468462,46.3260116582445,46.3828735382102,46.461914057841,46.548854827594,46.6268501275221,46.6882972672995,46.8252334570628,46.8333091669276,46.8952598567164,46.9571876465292,47.007858276464,47.0842323261294,47.2500686561455,47.4331130954694,47.5111923156076,47.5435829157846,51.0623168855353,51.4599876355413,51.4702758756166,51.5513877854321,51.5686111457242,51.6575012161962,51.6636810263659,51.6399993864819,51.6455573966642,51.5344848565705,51.520278926838,51.5322875970948,51.4688720674077,51.295833587518,51.2777786275585,51.2772827076283,51.3458862277179,51.3544654777856,51.3609008776923,51.3925170876693,51.4769439679528,51.488056178097,51.501098628103,51.4925003079501,51.5077781678344,51.5740966778326,51.6016883878827,51.5988883977986,51.6375122076957,51.6458320575773,51.6927795375384,51.7036094673994,51.7397232073788,51.7613906872552,51.7991294874456,51.7789306576088,51.7997589076653,51.8074951177866,51.8589477478474,51.862777707957,51.8833007779416,51.8763885478134,51.8898925777446,51.9071464478002,51.9070014976539,51.9388809177572,51.9697570780721,51.9901122981233,52.0059204080643,52.0080566381672,52.0183639481343,52.0316772482237,52.0291671781531,52.0488891581068,52.0519447282397,52.0705947882631,52.0777778582077,52.1069450383712,52.2241668684623,52.2097206084399,52.2322997983985,52.2463874785309,52.2583007785055,52.2616653385493,52.2541160584234,52.2886123683933,52.3145141585005,52.3600006085044,52.3611411985358,52.3713874784318,52.3780555684925,52.3913879384654,52.4171028085084,52.4227790784256,52.4425048784595,52.4405555684966,52.4749717685203,52.4788780184271,52.4897232084757,52.5027770984071,52.5200004584019,52.518115998349,52.5308837883619,52.5141677882409,52.5263900782658,52.5275001482137,52.5386123682501,52.5430908181616,52.5224990780815,52.5311126681093,52.5336112980595,52.5386123681416,52.5500106780287,52.5641288781179,52.5459022479644,52.5686225879925,52.566730498055,52.5889358481131,52.5919189480425,52.6058959981011,52.6063880880388,52.6166648881132,52.6197204580351,52.6425018280903,52.6491661080488,52.6294441180475,52.6218872079673,52.6420059179674,52.640029907862,52.6549987780228,52.679443358099,52.690917968038,52.6824989278714,52.7002792378302,52.700042717946,52.7166747979727,52.726665497911,52.7282714779842,52.7409057579162,52.7258338878662,52.7458343477649,52.7405166577163,52.7791671778466,52.7547073379098,52.7697143580022,52.779998777973,52.7663879380345,52.7813873281042,52.7758331280346,52.7891654980703,52.7969436580072,52.8261108380729,52.8046874979606,52.8153076179129,52.8389282180682,52.8491668680275,52.8614311181249,52.8505554181356,52.864723208144,52.8719444280708,52.8536987278683,52.8733329777972,52.8616943376986,52.8769607477985,52.8725013677156,52.8866538977228,52.8841667176662,52.9025268577243,52.9044456476289,52.9163894676174,52.957778927833,52.955276487955,52.9669456479461,52.9727783177979,52.9566650376221,52.984497067893,52.9877777077597,52.9983329778891,52.9961128177872,53.0180549577832,53.0041122377139,52.98916625775,52.9824829075696,52.9981079076195,52.9916992175071,53.0089111275818,52.9961128174767,53.0172729473999,53.029724117518,53.0383338874218,53.0367202775422,53.0499992375952,53.044998167536,53.0577773975939,53.0711059575032,53.0819435075214,53.0658340474018,53.0905570974989,53.0961112974517,53.1227760275263,53.1241683974535,53.1650009175964,53.1705779974526,53.2041664076591,53.2094459475437,53.220520017575,53.2208328176692,53.2411117576552,53.2355537375156,53.2625122076425,53.2750244074231,53.2821309631848,53.2710342373451,53.292171477429]},{"lng":[-61.5838946689643,-61.5868592044787,-61.5887792674175,-61.5838946689643],"lat":[50.1627775914425,50.1839750445132,50.1825256385164,50.1627775914425]}],[{"lng":[-57.2699613695135,-57.2655402359103,-57.2851816134418,-57.2699613695135],"lat":[51.4647146061285,51.4544912197528,51.4578977336621,51.4647146061285]}],[{"lng":[-57.6315784354719,-57.6183226092713,-57.643365573444,-57.6315784354719],"lat":[51.4247477681483,51.4036791962797,51.4109614409681,51.4247477681483]}],[{"lng":[-57.6762742652818,-57.6816757365137,-57.6934591164268,-57.6762742652818],"lat":[51.4083407213307,51.3902170981493,51.4075663312957,51.4083407213307]}],[{"lng":[-57.703369542384,-57.7255247995102,-57.7549698777069,-57.7505029791847,-57.726123757899,-57.703369542384],"lat":[51.4363901755956,51.4017489029638,51.4185907765873,51.4344065090552,51.4451448926533,51.4363901755956]}],[{"lng":[-57.787115624681,-57.7723797197887,-57.7939361364849,-57.787115624681],"lat":[51.3712846891576,51.3521425470713,51.3457910655214,51.3712846891576]}],[{"lng":[-57.8591090235991,-57.8405279745184,-57.8690155898141,-57.8591090235991],"lat":[51.3778840819264,51.3681795013061,51.3648797769868,51.3778840819264]}],[{"lng":[-58.3339125568017,-58.2978031401303,-58.2978031207433,-58.2816900857493,-58.3011332854346,-58.3066912435851,-58.3261344639022,-58.3444676436138,-58.327801472668,-58.3339125568017],"lat":[51.2700004401891,51.2530555618632,51.2336120518824,51.2388877872196,51.2174987708061,51.2377929589686,51.2330550927005,51.249168376883,51.2463874621547,51.2700004401891]}],[{"lng":[-58.3194664633603,-58.3087969010878,-58.356132775162,-58.3633005034131,-58.3194664633603],"lat":[51.2249984548462,51.2161102183061,51.1966666933077,51.2077865310837,51.2249984548462]}],[{"lng":[-58.3659402824777,-58.3652688889722,-58.3383565844388,-58.3433575578442,-58.379531807906,-58.3772468272964,-58.4037013679889,-58.4250214378749,-58.4077983202998,-58.4383536214124,-58.3659402824777],"lat":[51.25657269022,51.24290082044,51.2413902088124,51.2163886772663,51.2130546261318,51.1994437868393,51.1912116589492,51.2077788827204,51.2119445377195,51.2150000989006,51.25657269022]}],[{"lng":[-58.3472447169828,-58.3550227627132,-58.3639108955421,-58.3472447169828],"lat":[51.2630538659999,51.2502784435917,51.2608337108383,51.2630538659999]}],[{"lng":[-58.4016911410147,-58.3761330148981,-58.4044681894245,-58.4016911410147],"lat":[51.2819442294401,51.2644462271046,51.2683333986351,51.2819442294401]}],[{"lng":[-58.5127637834502,-58.5344636476281,-58.5483527926895,-58.556130844487,-58.578908125509,-58.5290392539341,-58.5244654830465,-58.5250224392946,-58.4178003838257,-58.417243419136,-58.40057724866,-58.39057905806,-58.4033543040831,-58.4000241024443,-58.4311325090062,-58.4178003464663,-58.4339095860375,-58.4414701957136,-58.4672457597016,-58.4822449370797,-58.4987281489474,-58.4939101198588,-58.5577978398236,-58.5127637834502],"lat":[51.2190878989508,51.2227782429898,51.2397231294471,51.2183341175262,51.2313880019015,51.2641066743299,51.2519454255227,51.2675017553609,51.2791671347279,51.2550010249146,51.2627791997883,51.2511100427709,51.2566680489752,51.2372207199792,51.2243155909468,51.2388877447707,51.2408332301344,51.2225608280083,51.2266654408126,51.2138900167357,51.2166985160972,51.2116660436015,51.214443127116,51.2190878989508]}],[{"lng":[-58.4254219432143,-58.4879477523902,-58.533910445766,-58.5316864903372,-58.5772410393423,-58.5473303968898,-58.4254219432143],"lat":[51.1762275226459,51.1254424353224,51.1313895432618,51.1150016038571,51.1277770124576,51.147655409832,51.1762275226459]}],[{"lng":[-58.4561336689003,-58.4416876037291,-58.4594676514941,-58.4811310346734,-58.4561336689003],"lat":[51.1925010139311,51.1908339979867,51.1788901730229,51.1833342870776,51.1925010139311]}],[{"lng":[-58.4527997669949,-58.4205774381418,-58.5000212445883,-58.4527997669949],"lat":[51.3058318547334,51.2822227039202,51.2747229818992,51.3058318547334]}],[{"lng":[-58.546132641738,-58.518911289765,-58.5750209577446,-58.546132641738],"lat":[51.2052764100508,51.1763877170603,51.1874999129078,51.2052764100508]}],[{"lng":[-58.5450187431825,-58.5270670009151,-58.5383545340262,-58.5616887525278,-58.5450187431825],"lat":[51.178611680369,51.1734236949543,51.1611098420832,51.1702765562115,51.178611680369]}],[{"lng":[-58.5576108361544,-58.577862797074,-58.5994651685179,-58.5794649803728,-58.5576108361544],"lat":[51.1134947873128,51.0987776823545,51.1130560871397,51.1266669319187,51.1134947873128]}],[{"lng":[-58.6172414657267,-58.5694630034095,-58.548932549609,-58.5939072368561,-58.6033523262057,-58.6272434517592,-58.6494637974593,-58.6361506812147,-58.6605758733791,-58.6172414657267],"lat":[51.1463889129089,51.1699980842941,51.1514815494231,51.123889828445,51.144443416168,51.1086119606586,51.1163901256032,51.1311568185839,51.1163901231303,51.1463889129089]}],[{"lng":[-58.6039092315149,-58.5927971415154,-58.6122404057556,-58.6055762393468,-58.6039092315149],"lat":[51.0988883061118,51.0888899887697,51.0836104341808,51.109165095701,51.0988883061118]}],[{"lng":[-58.6524354104799,-58.6322406313314,-58.6644629795996,-58.6524354104799],"lat":[51.1150969449409,51.097221269533,51.0825003523325,51.1150969449409]}],[{"lng":[-58.678775573549,-58.6935916563936,-58.760367152943,-58.7458829061632,-58.7555721151792,-58.7733522235296,-58.7881034676317,-58.8055746201519,-58.678775573549],"lat":[51.1138075691538,51.0736311760478,51.0719259924117,51.0564345053064,51.0424994134115,51.0550001799188,51.0136869971821,51.0630568138098,51.1138075691538]}],[{"lng":[-58.7366857607072,-58.7244636154142,-58.7494610263039,-58.7366857607072],"lat":[51.0422209471799,51.0472220196551,51.0313872046433,51.0422209471799]}],[{"lng":[-58.8599638138975,-58.8639043458523,-58.8413177621274,-58.8568243192782,-58.8811275140894,-58.8965921277564,-58.8599638138975],"lat":[50.831065984604,50.8080557339861,50.8161886679047,50.7776564152923,50.793056341099,50.7855566385464,50.831065984604]}],[{"lng":[-58.9081163158589,-58.8945590110507,-58.9372374116074,-58.9081163158589],"lat":[50.9413298062665,50.9327963285056,50.9341657916502,50.9413298062665]}],[{"lng":[-59.2409644608727,-59.2215058548043,-59.2272354631371,-59.2409644608727],"lat":[50.7149732538862,50.711986345849,50.6894453053338,50.7149732538862]}],[{"lng":[-59.3708156567575,-59.3260543591999,-59.3367880643656,-59.3411375379411,-59.3770526050819,-59.3642734826735,-59.3915750777277,-59.3708156567575],"lat":[50.644595892624,50.6407277962919,50.6144305752598,50.6201894650895,50.5801923623406,50.5956342532922,50.6292608110781,50.644595892624]}],[{"lng":[-59.334207279123,-59.2996987920516,-59.3047188764733,-59.2927484577734,-59.3527912943454,-59.3663753387635,-59.3300063188868,-59.3477178259771,-59.3294570228937,-59.334207279123],"lat":[50.6110128582594,50.5845334887675,50.5569341584116,50.5643880794576,50.5216672744551,50.5424497432987,50.5865247561276,50.601466924607,50.6047208661159,50.6110128582594]}],[{"lng":[-59.2761242058944,-59.2752544540244,-59.2927904838216,-59.2761242058944],"lat":[50.6775014806258,50.660918990757,50.6686094191246,50.6775014806258]}],[{"lng":[-59.3011064526867,-59.3109368389179,-59.3135880380551,-59.3011064526867],"lat":[50.6627118983967,50.6554563375512,50.6753080272598,50.6627118983967]}],[{"lng":[-59.4409788214347,-59.4248437068134,-59.4611240879456,-59.4409788214347],"lat":[50.5871656176711,50.5830726580279,50.5794446763522,50.5871656176711]}],[{"lng":[-59.4198796353555,-59.4094543953702,-59.4188995015857,-59.4453962008798,-59.4239005386218,-59.4198796353555],"lat":[50.581812853173,50.5791661999142,50.5550420593127,50.5717504274207,50.5647237589267,50.581812853173]}],[{"lng":[-59.4766764608619,-59.478900406234,-59.4966767615723,-59.4766764608619],"lat":[50.5113904755749,50.4938886354934,50.5024984143683,50.5113904755749]}],[{"lng":[-59.8016406958579,-59.7838986423782,-59.7916767591237,-59.8420533456401,-59.8393754481369,-59.7961208554235,-59.8226442920707,-59.8022319759398,-59.8016406958579],"lat":[50.4054563119932,50.4055555024431,50.3891675622972,50.3740003211935,50.3921697211871,50.3966672521594,50.4075009914952,50.4147222119477,50.4054563119932]}],[{"lng":[-59.9077839848756,-59.9027142796506,-59.9255451126914,-59.9077839848756],"lat":[50.2747228204041,50.265228040511,50.2637670102058,50.2747228204041]}],[{"lng":[-60.0088080538922,-60.0122298088059,-60.0432469448768,-60.0513798468216,-60.0088080538922],"lat":[50.2269437396001,50.2055547296589,50.1928670495899,50.2028195895373,50.2269437396001]}],[{"lng":[-60.0740161410504,-60.0648151323988,-60.1222298567593,-60.1144517295864,-60.0740161410504],"lat":[50.1974980895371,50.1828572795923,50.1761090797058,50.1919438896228,50.1974980895371]}],[{"lng":[-60.4933372735215,-60.4877830905388,-60.5180945604081,-60.4933372735215],"lat":[50.2219426755397,50.204246295446,50.2270658162064,50.2219426755397]}],[{"lng":[-60.5110641026032,-60.5333380257933,-60.5333380287092,-60.5110641026032],"lat":[50.2086904160663,50.1966664967352,50.2088887866974,50.2086904160663]}],[{"lng":[-61.9116837070275,-61.9340835750081,-61.8955055887395,-61.8295915145029,-61.8358666848207,-61.7756975514697,-61.8380067311634,-61.8812271945857,-61.8605057820041,-61.8949791585677,-61.9115005865861,-61.8832833119535,-62.0065016889419,-62.0208983438316,-61.9610000524116,-61.9582839915867,-61.9351593322003,-61.8085611440547,-61.7125567990642,-61.5468771410732,-61.4328026316009,-61.39167264596,-61.3876176218996,-61.4522155921731,-61.4649490256596,-61.5313704175414,-61.5617315384624,-61.5232222337136,-61.4916175315027,-61.4799216800974,-61.4840720674142,-61.4452270785732,-61.5391180620936,-61.5166037523331,-61.5355856600423,-61.5889760686978,-61.8648087743444,-61.8693940363244,-61.8396012863962,-61.8527238264805,-61.8800751673528,-61.9116837070275],"lat":[47.3496702610852,47.3079222124118,47.2492750606775,47.2393186973424,47.2260321077221,47.2697142044503,47.2176092578688,47.2208860800674,47.2248304789889,47.2398794606907,47.2299192815804,47.2212179601714,47.2156141765092,47.2379759271557,47.3173216337573,47.3851088033407,47.405452582065,47.4576071153133,47.5315283100489,47.6243284411769,47.6347044154215,47.6235311634529,47.6109884433169,47.5997579765528,47.5689962573342,47.5424497807804,47.5567777822426,47.5623663202765,47.590717148558,47.5756834180467,47.613097968074,47.6303861760576,47.6070516808657,47.6237180896537,47.6239012006088,47.5998724234232,47.4196775783756,47.3984411587015,47.4020994571505,47.3773191879321,47.3443830894801,47.3496702610852]}],[{"lng":[-61.445673419484,-61.4362778356852,-61.4710296611837,-61.5115264198947,-61.445673419484],"lat":[47.8027113152581,47.7954290548239,47.7813871566265,47.7806699986685,47.8027113152581]}],[{"lng":[-61.6885509832853,-61.7735689663453,-61.8363053909365,-61.7667139704506,-61.7674578325005,-61.7125262744062,-61.6885509832853],"lat":[47.5115239498722,47.3899534538067,47.4030760169766,47.4342230232602,47.4655073531618,47.4860838202486,47.5115239498722]}],[{"lng":[-61.6822366550866,-61.6577778334156,-61.5797483292206,-61.6822366550866],"lat":[47.518218502869,47.5441282571845,47.5601270931385,47.518218502869]}],[{"lng":[-61.685003254275,-61.692174871841,-61.7133349707695,-61.7136134416833,-61.685003254275],"lat":[47.2841680797627,47.2676618902037,47.2691648812722,47.2819441112275,47.2841680797627]}],[{"lng":[-62.1772246273132,-62.1427779485366,-62.0869461046609,-62.0405556341496,-61.9769990463232,-61.9678361467564,-61.8970697958418,-61.8761004334903,-61.833787874543,-61.8442401260413,-61.8036136664663,-61.8048992123667,-61.7206670081799,-61.6991368881347,-61.6687185368252,-61.7138921087198,-61.7005559388415,-61.770101579225,-61.8259601025086,-61.9078500964469,-61.9585397256828,-62.0566688691285,-62.064725498285,-62.0861144829525,-62.0661140464676,-62.2094854573127,-62.2505849612127,-62.5361109559623,-62.5616694131771,-62.7580574967245,-63.0997244561415,-63.2352796442887,-63.3052793109623,-63.3300252386468,-63.4261097855904,-63.5962528463824,-63.5749859150109,-63.6145519407238,-63.6341671147212,-63.8266604906339,-63.9820635751595,-64.2679750651133,-64.374542433781,-64.3971406892209,-64.3804933521829,-64.4161760253314,-64.4560243482674,-64.5220185966637,-64.519691637237,-64.462555107822,-64.1491320151396,-64.0769427030168,-63.9880068770207,-63.8701327712373,-63.3799176490667,-63.0814177453473,-63.0122764070674,-63.0021407627705,-62.9411132722411,-62.9158332881306,-62.8533333312933,-62.7608346173737,-62.7002801646411,-62.6430559293355,-62.5133868508854,-62.4419681556499,-62.4605571570876,-62.4150020897496,-62.415280558561,-62.3944446985758,-62.3488896352007,-62.3183339355307,-62.2529996853343,-62.2455572212584,-62.1772246273132],"lat":[49.3966673385727,49.4038885566981,49.3772238237388,49.3866651911978,49.3676107777857,49.3445700073446,49.3526037534667,49.3252981523953,49.3139952001202,49.2818983407767,49.2774999885784,49.2551344187108,49.1984976043063,49.1516798232838,49.1393888716829,49.0977781542436,49.084445783566,49.087215257311,49.0630491604056,49.0816038447893,49.067241517576,49.0791662628495,49.0694426033098,49.0838888644249,49.0683325233876,49.072955951069,49.0625990432822,49.1425017080054,49.1336096493273,49.1580542390921,49.2319449450656,49.2802771610022,49.3355559439412,49.3213385650105,49.3574980889697,49.3911132057288,49.4086112248804,49.4498862463741,49.5125006870641,49.6368979740977,49.6973342294686,49.7565001885784,49.8302917016776,49.8198318023325,49.7925071218714,49.7995910128856,49.8395537839817,49.8618621357802,49.879188495708,49.9133910641229,49.9523200447707,49.9262618424598,49.9192084695029,49.876514365449,49.8266142965177,49.7671011934884,49.7517737303159,49.7332800798677,49.7313879869984,49.705001725833,49.7016676828344,49.6577757683772,49.659721255364,49.6205557625499,49.5882223759714,49.5459974923341,49.5330542233293,49.5094450609917,49.4941672010362,49.4841650599613,49.4908331475436,49.4666670459725,49.4551657225126,49.4211119221878,49.3966673385727]}],[{"lng":[-63.0656325620661,-63.0486533520313,-63.0632064126977,-63.0872237284113,-63.0656325620661],"lat":[50.2577857021974,50.2415236414275,50.2331580121108,50.2447279932064,50.2577857021974]}],[{"lng":[-63.1327101508459,-63.1255576053571,-63.0992171305134,-63.1087042744064,-63.1988913025832,-63.1957861349119,-63.1581846860339,-63.1372229386449,-63.1327101508459],"lat":[50.2297934552994,50.2394446349647,50.2252539637768,50.2122801742241,50.2099989882949,50.2293242481375,50.2271346064542,50.2427786854911,50.2297934552994]}],[{"lng":[-63.3422246578638,-63.2830587371612,-63.3499990035461,-63.3422246578638],"lat":[50.2158316745398,50.1963156820105,50.1977767048875,50.2158316745398]}],[{"lng":[-63.6272854129534,-63.5919804068172,-63.604999961199,-63.6605571899098,-63.6383518488066,-63.6272854129534],"lat":[50.2294005561519,50.2252349147747,50.2061118353007,50.216945577447,50.2360800965761,50.2294005561519]}],[{"lng":[-63.7392734412191,-63.7294467842923,-63.7466663180406,-63.7695964605378,-63.7392734412191],"lat":[50.2239646204236,50.2026976800715,50.1949996207205,50.2200545615528,50.2239646204236]}],[{"lng":[-63.8070758478706,-63.7805560867551,-63.8283351503641,-63.8405536168171,-63.8070758478706],"lat":[50.2236518229253,50.2191657319575,50.1983336737145,50.2227134041383,50.2236518229253]}],[{"lng":[-64.1606829026084,-64.1430895163822,-64.167793490679,-64.1606829026084],"lat":[48.5080604063316,48.493331855801,48.483268686576,48.5080604063316]}],[{"lng":[-63.9022219976656,-63.8509181533065,-63.861111019592,-63.910000164381,-63.924537970202,-63.9261897301333,-63.9022219976656],"lat":[50.2441672663178,50.2138900145165,50.1886100148984,50.1914367066256,50.2055625271243,50.2313117371651,50.2441672663178]}],[{"lng":[-71.5101394699961,-72.3084182699997,-73.05492401,-74.15013123,-74.6375274700002,-74.5353469800001,-74.4688949600001,-74.4056625400001,-74.4069366500001,-74.3513107300001,-74.3361434900001,-74.22350311,-74.16442108,-74.15733337,-74.17888641,-74.13414764,-73.92343903,-73.76531982,-73.51839447,-73.50715637,-73.43965149,-73.41176605,-73.27006531,-73.15090942,-72.967659,-72.93516541,-72.74355316,-72.4872283899999,-72.2883529699997,-72.2296676599996,-72.1942138699996,-72.1527786299995,-71.935897829999,-71.8484878499987,-71.6534194899977,-71.4890899699964,-71.3917617799955,-71.2613906899939,-71.2113876299932,-71.1630554199925,-70.9908370999894,-70.881111149987,-70.840835569986,-70.7952804599849,-70.7680587799842,-70.7516632099837,-70.62139129998,-70.5500030499777,-70.5011138899761,-70.3697204599718,-70.3152923599699,-70.1955566399662,-70.1094436599631,-70.0601654099616,-70.0458297699615,-70.0255355799608,-70.0611114499621,-70.025001529961,-70.0233306899612,-69.9891662599603,-69.9732818599597,-69.9324493399589,-69.9199523899584,-69.9004363999579,-69.8557968099574,-69.7786636399563,-69.7454910299566,-69.6381607099572,-69.5564256426787,-69.5681304899599,-69.4974975599636,-69.5150604199631,-69.3886108399717,-69.3486099199747,-69.2632904099858,-69.2158355699924,-69.2027740499953,-69.1341629000082,-69.1051254300155,-68.9538879400602,-68.8061142001223,-68.8305587801112,-68.8194427501171,-68.7841644301333,-68.7522201501515,-68.7511138901508,-68.5791702302709,-68.5388870203045,-68.5175018303267,-68.4619445803826,-68.4741668703714,-68.3522186305113,-68.1650238008071,-68.1252746608791,-68.0844421409626,-68.0616684010153,-68.0336074810786,-68.01860809111,-67.9616699212519,-67.8338851916297,-67.5988845825497,-67.5744171126728,-67.4801635731586,-67.3691635138379,-67.1600036654358,-66.6474990920005,-66.4797210852292,-66.409317036831,-66.169998193304,-65.9631958305108,-65.8886108735148,-65.8354645158607,-65.8024978973357,-65.7572250795391,-65.7297897708653,-65.6769409636586,-65.5672226498864,-65.3247223563947,-65.276878430084,-65.0481492008143,-64.8531724123721,-64.7446366659821,-64.6085511754745,-64.4746400568636,-64.2129747659403,-64.2005464784213,-64.2198869935543,-64.1631243053526,-64.2997438471115,-64.5908357162489,-64.4731523248976,-64.5008775400515,-64.5752793880709,-64.5725022883864,-64.497291720556,-64.4566041676728,-64.4061128465885,-64.3790590212965,-64.4341661210213,-64.3353502292018,-64.1676180226654,-64.275300606773,-64.2890398059529,-64.2043077732383,-64.2176668400346,-64.3069307314229,-64.3285448068005,-64.3169481288171,-64.4697190910143,-64.5558320568299,-64.5961076207344,-64.5972215104049,-64.6745835691102,-64.6920090965625,-64.7036134051265,-64.7302781415417,-64.7300034814412,-64.6939164462937,-64.7105561539446,-64.6952744759208,-64.741111879662,-64.7736130949285,-64.9364014659019,-64.9727784122387,-64.9775010115025,-64.9488068544226,-65.1502915147649,-65.133613666165,-65.167587353178,-65.3278198805782,-65.4527741022553,-65.4708328713024,-65.490280200111,-65.4828415405228,-65.5350494875627,-65.8145828538247,-65.8554077422011,-65.8391647629037,-65.9004822104476,-65.9019470505476,-65.9095001501703,-65.9691696479181,-66.0462265250992,-66.0922241435875,-66.1247253625798,-66.1005554433399,-66.1277771225252,-66.1242752326036,-66.1362686422819,-66.1725692912375,-66.2761764684646,-66.2991867278829,-66.2693329086136,-66.2957611279005,-66.3576889165041,-66.4405670347257,-66.4625702042581,-66.4735489040547,-66.5445861926518,-66.5350341928041,-66.5797500719912,-66.6478119008703,-66.6997604500296,-66.7246017596754,-66.7869491687894,-66.8505935779885,-66.9212722871525,-66.964614876653,-66.9701156665778,-66.9447326768363,-66.9573974666928,-67.0592804057226,-67.2108459544349,-67.2877578739075,-67.304847723789,-67.3486404435189,-67.3366622935817,-67.3754196233364,-67.3933334432438,-67.4832077027788,-67.594657902274,-67.6138076822119,-67.5980529822845,-68.1226043707818,-68.1227951007678,-68.3823318504034,-68.3834991503611,-68.5692749001916,-68.7996292100647,-69.0505294799913,-69.0439987199978,-69.2218170199715,-69.9970855699539,-70.0571594199545,-70.0952835099563,-70.1291198699576,-70.1953430199607,-70.2920074499645,-70.2386169399617,-70.250839229962,-70.2830123899636,-70.2995910599643,-70.3085327099647,-70.2901000999638,-70.3142394999648,-70.3144302399645,-70.2754821799626,-70.2404785199608,-70.2619628899616,-70.3455123899655,-70.4187774699687,-70.3964767499675,-70.5613098099745,-70.685340879979,-70.7184371899801,-70.7157974199799,-70.6316909799767,-70.6392593399769,-70.7140121499797,-70.7545165999811,-70.7961196899825,-70.8233261099833,-70.8066482499827,-70.8352813699835,-70.8422775299836,-70.8686904899844,-70.956588749987,-70.9790573099875,-71.0096359299883,-71.0085830699882,-71.0996932999902,-71.1623306299914,-71.2285079999926,-71.2719116199933,-71.2997436499937,-71.3602600099945,-71.3784561199947,-71.4367217999954,-71.4103317299951,-71.4284286499953,-71.489051819996,-71.5101394699961],"lat":[45.0155639599947,45.0036849999997,45.01567078,44.99119568,44.9993286099998,45.0364532499999,45.0441665599999,45.0979003899999,45.1252975499999,45.14096832,45.16035843,45.17531586,45.20027924,45.22175598,45.23123169,45.28553009,45.30492401,45.36883163,45.42120361,45.5203743,45.59692001,45.6966095,45.80437088,46.04571533,46.10179901,46.12810135,46.16528702,46.3131256099999,46.3776245099997,46.4260330199996,46.5175971999996,46.5426788299994,46.5787925699987,46.6233367899982,46.6168136599964,46.692398069994,46.7058677699921,46.7482757599887,46.7741661099871,46.8325004599853,46.8352775599775,46.8927764899709,46.8841667199681,46.9158325199647,46.9055557299625,46.9272232099611,46.9824981699486,46.9861106899405,47.0096664399343,47.1343879699148,47.1614379899055,47.3071670498817,47.3335533098619,47.3714370698493,47.4230003398453,47.4265670798398,47.4271659898494,47.4566116298396,47.486053469839,47.5020675698292,47.4947662398246,47.5344238298119,47.522338869808,47.5280761698016,47.5816078197861,47.6204872097573,47.6731452897438,47.7392883296964,47.8468790920817,47.8471641496611,47.9300003096224,47.9390525796321,48.0227775595558,48.0130538895294,48.0898704494678,48.0963897694311,48.1237220794203,48.1527786293624,48.1926917993362,48.2900009191861,48.3472213690138,48.351112369044,48.3658332790301,48.3516654989859,48.3761100789438,48.3566665589427,48.4430541986909,48.4455566386258,48.4813880885892,48.5091667184921,48.5194435085135,48.5488891582846,48.6426124578752,48.6319427477798,48.6455573976771,48.6733322076174,48.6813888475431,48.6655540475035,48.6847228973447,48.7505569469567,48.8194503761244,48.8460617060267,48.8560562056376,48.8986129751374,48.9422225940703,49.1144447206202,49.1297225891961,49.1626129085442,49.2077789161007,49.2281226936719,49.2202758627212,49.2338142220122,49.2231025515661,49.2405547909304,49.229206070543,49.248889899767,49.2572211980882,49.2526664439987,49.2337913231332,49.2246436786472,49.1874389244046,49.1348342518813,49.1114959285053,49.0542830949963,48.8747214775878,48.8557738772254,48.8069457478587,48.7483062161718,48.8260497502242,48.8958320181939,48.8373031151166,48.8237723858765,48.8311118678367,48.8183326277737,48.8140525357874,48.8282203246718,48.8180541532808,48.7912215725403,48.7708320140993,48.7534865813221,48.6258925864288,48.6154454874933,48.5533751901526,48.5256156376506,48.4999465380801,48.4724463907498,48.4367904214077,48.4180564410908,48.4036101853745,48.378055527678,48.3824996487115,48.364723168754,48.3458289707056,48.327571831146,48.3438872914159,48.3393935820618,48.3224982820676,48.3264464911933,48.3136100416065,48.3038901912433,48.281387292363,48.2056388531856,48.1762008368671,48.1988906576311,48.1594428677566,48.1509437271509,48.0657958713107,48.0586127909912,48.0431136816568,48.002231574621,48.0051154867434,48.0475005870186,48.0452766173344,48.035415627219,48.068405128036,48.122257212076,48.1526985026055,48.1652793723877,48.1698455631814,48.2337226731791,48.1992187332863,48.1880569340324,48.1124915949744,48.0927772355078,48.0851096958738,48.0980338956003,48.1041679259022,48.0904426458672,48.1090736259944,48.113670336388,48.1126098474666,48.1057739176981,48.0981635874011,48.0642051576743,48.1079482882668,48.1231689390334,48.108776079234,48.1207771193289,48.0765075599512,48.0585937398741,48.0442771802502,48.0516395507954,48.0101737912031,48.0188064513879,47.9918746818458,48.0012054322868,47.9846038727576,47.9406776330393,47.9158897330771,47.9048538129193,47.8921089130011,47.9358253436116,47.8754806444536,47.8988799948389,47.8857192949234,47.8958969051304,47.8727531350765,47.8444023052591,47.8683052053381,47.9030990557258,47.9249725261712,47.9793434062399,47.9998931861784,47.9998435977991,47.9163207978033,47.9161148083631,47.5531997683778,47.4271240187021,47.3489417990259,47.3001708992987,47.4257736192905,47.4576415994433,46.6958312998347,46.4146804798521,46.4081153898617,46.3693542498698,46.3433990498845,46.1909408599037,46.1456413298938,46.0979118298964,46.0998992899023,46.0835838299054,46.063480379907,46.0613708499037,46.0208015399081,45.9664916999083,45.9709052999013,45.9483032198948,45.8895492598991,45.8507614099138,45.7932891799252,45.7262802099221,45.6627311699439,45.5684089699568,45.5159034699598,45.4875717199597,45.4232521099519,45.3812255899527,45.3925208999596,45.427650449963,45.4293823199662,45.4029693599682,45.3211402899671,45.2945022599692,45.2429923999697,45.2309760999714,45.3426094099766,45.3349609399779,45.3461799599794,45.3244705199794,45.3023185699835,45.2468604999859,45.2497405999882,45.2961845399894,45.2965087899902,45.2685813899917,45.2352485699921,45.2371101399934,45.1937904399928,45.1270484899932,45.0753669699943,45.0155639599947]},{"lng":[-64.2753199951939,-64.3225023300262,-64.2966692049571,-64.2938921051608,-64.2753199951939],"lat":[48.6154435967855,48.6108321610752,48.5969428503362,48.5680541502813,48.6154435967855]}],[{"lng":[-63.9788897643665,-64.0011141780602,-63.9866564823325,-63.9788897643665],"lat":[50.2180556689909,50.2205542897446,50.2305411692478,50.2180556689909]}],[{"lng":[-64.0022204381762,-63.985870652743,-64.0049975371367,-64.0288928005825,-64.0022204381762],"lat":[50.248054439765,50.2422522892137,50.231666499869,50.239166200668,50.248054439765]}],[{"lng":[-64.0077746371728,-63.987778002757,-64.0372012089289,-64.0077746371728],"lat":[50.2844428399306,50.2777785692567,50.2811240509206,50.2844428399306]}],[{"lng":[-66.2910614213241,-66.272506731823,-66.3038864309106,-66.2910614213241],"lat":[50.1580200072306,50.1373443470414,50.1236114373664,50.1580200072306]}],[{"lng":[-66.3680725292635,-66.3572235295027,-66.3777771189806,-66.3680725292635],"lat":[50.1768684280009,50.1463889978981,50.1536102180983,50.1768684280009]}],[{"lng":[-66.4121933180604,-66.3816681088092,-66.3903122085796,-66.4121933180604],"lat":[50.111457808434,50.1022224281423,50.0941314582268,50.111457808434]}],[{"lng":[-65.1815873471813,-65.1561128006933,-65.1922227256231,-65.2041245740463,-65.1815873471813],"lat":[59.9725188950009,59.9522208945477,59.950553865156,59.9646262853694,59.9725188950009]}],[{"lng":[-65.1716920184744,-65.1768952677052,-65.1976167048641,-65.1716920184744],"lat":[59.9490966548089,59.938941934886,59.9459190152413,59.9490966548089]}],[{"lng":[-65.4055558272038,-65.4197159756319,-65.4346924740575,-65.4055558272038],"lat":[59.3322219579935,59.3216209182013,59.3312988084367,59.3322219579935]}],[{"lng":[-65.4552918421419,-65.4316712346166,-65.456909271938,-65.4552918421419],"lat":[59.4163932588163,59.4095000984563,59.4044799588305,59.4163932588163]}],[{"lng":[-65.4984055479747,-65.4672242111212,-65.5066681771276,-65.4984055479747],"lat":[59.5006561095188,59.4910888490548,59.4925002896317,59.5006561095188]}],[{"lng":[-65.5789185400749,-65.5083313867087,-65.556518632078,-65.5789185400749],"lat":[59.4408340306153,59.4013900595832,59.4088744902775,59.4408340306153]}],[{"lng":[-65.5933228277434,-65.5966645074143,-65.6194458754474,-65.5933228277434],"lat":[59.0441665405254,59.0325012005639,59.0502776908929,59.0441665405254]}],[{"lng":[-65.5455551933555,-65.527687155024,-65.5531006725932,-65.5455551933555],"lat":[59.4983329601918,59.4786414899226,59.4815063302852,59.4983329601918]}],[{"lng":[-65.6111145768396,-65.5838852691939,-65.6268158754016,-65.6111145768396],"lat":[59.3136100609658,59.2758331105614,59.2912216011643,59.3136100609658]}],[{"lng":[-65.7233353369139,-65.6939087592068,-65.7594452540966,-65.7233353369139],"lat":[59.0800018123041,59.0574951019037,59.0627784527574,59.0800018123041]}],[{"lng":[-65.7714005134591,-65.7433319755371,-65.7844238924139,-65.8171692500606,-65.7714005134591],"lat":[59.1859779229865,59.1608886526143,59.1489257631266,59.1592864835385,59.1859779229865]}],[{"lng":[-65.7850113524416,-65.8038864710596,-65.778076232973,-65.7850113524416],"lat":[59.1832046331554,59.1817359733888,59.191711413074,59.1832046331554]}],[{"lng":[-65.8263931891314,-65.8266678490848,-65.837829648353,-65.8263931891314],"lat":[59.0206909035681,59.0069427335633,59.031127913714,59.0206909035681]}],[{"lng":[-65.8004685014274,-65.7949982317941,-65.8188858600826,-65.8004685014274],"lat":[59.2408408933833,59.2249984533057,59.2316665436037,59.2408408933833]}],[{"lng":[-65.871826226161,-65.834999138657,-65.8633347166942,-65.871826226161],"lat":[59.0985107241626,59.0855560137118,59.0777778440492,59.0985107241626]}],[{"lng":[-65.8894424948205,-65.867225706233,-65.9336700919352,-65.8894424948205],"lat":[59.0072212043193,58.9733314340355,58.9804191448204,59.0072212043193]}],[{"lng":[-65.9052811238286,-65.8933716345866,-65.9277802023439,-65.9172211230851,-65.9052811238286],"lat":[59.0291786045178,59.0200614743729,59.0022239547644,59.0422210546642,59.0291786045178]}],[{"lng":[-65.95770269044,-65.9369736217015,-65.9766693592692,-65.9822235589115,-66.0532837348849,-65.95770269044],"lat":[58.9746703950922,58.96486280485,58.9594459352984,58.9433326553523,58.9644775261417,58.9746703950922]}],[{"lng":[-66.0738907335896,-66.0599976043283,-66.0905533226814,-66.1088867617439,-66.0850220130116,-66.0738907335896],"lat":[58.8483314363052,58.8411254761534,58.8322219664737,58.8436126566709,58.8580932464277,58.8483314363052]}],[{"lng":[-66.1181335815848,-66.0883331731337,-66.119583171495,-66.1181335815848],"lat":[59.0642928968702,59.0591659365592,59.05347059688,59.0642928968702]}],[{"lng":[-66.0960083427201,-66.1050034022228,-66.1390991604777,-66.0960083427201],"lat":[59.0536918466366,59.0355567767212,59.0338744970697,59.0536918466366]}],[{"lng":[-67.6077880951419,-67.621109015045,-67.55106354562,-67.559997565547,-67.4989013760983,-67.5122222959566,-67.5777740553749,-67.6383361848886,-67.6761093146104,-67.6721649246437,-67.6244430550084,-67.6522216847978,-67.6077880951419],"lat":[58.3447265564028,58.3869438164554,58.3619232161953,58.3795165962311,58.3947143559982,58.3222236560413,58.2881011962866,58.2841682365051,58.3061103766386,58.3280677766274,58.3391685464619,58.3549995365614,58.3447265564028]}],[{"lng":[-68.8407592801101,-68.8497238201052,-68.8905563400878,-68.8407592801101],"lat":[48.4119758590554,48.3955573990664,48.3858337391145,48.4119758590554]}],[{"lng":[-67.6111145051284,-67.6077728351521,-67.6261138950073,-67.6111145051284],"lat":[58.4044456464216,58.390556336408,58.394721976474,58.4044456464216]}],[{"lng":[-67.8911132832756,-67.9000015332241,-67.9183197031282,-67.8911132832756],"lat":[58.4227790773197,58.3963890073425,58.3983650173938,58.4227790773197]}],[{"lng":[-68.3703155515658,-68.2312469520176,-68.1395797723641,-67.9979171830004,-67.9598999031901,-67.8255157539624,-67.8546829237707,-67.957817083176,-67.9567718531774,-68.0604171826728,-68.1208343524094,-68.1640625022375,-68.204689032083,-68.3458328216228,-68.4161453214309,-68.4411468513704,-68.3703155515658],"lat":[60.391143798534,60.5875015282959,60.597915648113,60.5854148877959,60.5401039076995,60.4765624973412,60.4088516174136,60.3307304376735,60.2963561976675,60.2958335879145,60.2625007580455,60.2619781481369,60.227603908217,60.1958351084795,60.2109374985996,60.2630195586431,60.391143798534]}],[{"lng":[-69.3898467999725,-69.4937591599648,-69.4246063199699,-69.3898467999725],"lat":[48.0620689395563,47.9864273096198,48.0516776995783,48.0620689395563]}],[{"lng":[-69.703422549959,-69.7751541099585,-69.6862716699594,-69.703422549959],"lat":[47.876781459725,47.7982673597551,47.9028396597173,47.876781459725]}],[{"lng":[-68.9525375404785,-68.897087100535,-68.9593048104715,-68.9525375404785],"lat":[59.0508918792468,59.003894809189,59.0375442492531,59.0508918792468]}],[{"lng":[-69.058288570382,-69.0327758804036,-68.9813385004495,-68.9784545904516,-69.0225067104112,-69.0650024403761,-69.0522232103861,-69.0777740503656,-69.0652694703754,-69.1020736703466,-69.058288570382],"lat":[59.0097045893446,59.021389009322,59.0094184892738,58.9839858992704,58.9661827093113,58.9872207593499,58.9705543493383,58.9755554193608,58.9665946993497,58.9577789293811,59.0097045893446]}],[{"lng":[-68.9867019704471,-68.9675903304648,-68.9891967804446,-68.9929809604408,-68.9867019704471],"lat":[59.1065826392813,59.0885009792625,59.1007080092836,59.0919189492869,59.1065826392813]}],[{"lng":[-69.0633316003795,-69.0494461103907,-69.0699996903739,-69.0750122103704,-69.0633316003795],"lat":[59.0791664093506,59.0699996893381,59.0663871793561,59.0894775393609,59.0791664093506]}],[{"lng":[-69.3084716802186,-69.3497161901987,-69.2733154302389,-69.3477401702022,-69.3289184602115,-69.2961120602279,-69.2500000002534,-69.1810913102949,-69.1647644003059,-69.1828002902927,-69.1638870203051,-69.1527099603129,-69.1122421079856,-69.1511077903145,-69.1222229003351,-69.1255569503331,-69.0927124003574,-69.0810852103657,-69.1188964803372,-69.0822143603642,-69.109649853222,-69.090011600358,-69.1555557303101,-69.1503906203131,-69.2033081102782,-69.2244873002646,-69.2736129802373,-69.3084716802186],"lat":[58.9506835895355,58.9482879595621,59.0741653395138,59.1344680795637,59.153930659552,59.1408348095304,59.1652831994991,59.134704589448,59.1461029094357,59.0753173794482,59.0708351094335,59.086730959425,59.0689479044337,59.1091651894242,59.1186103794012,59.1397209194044,59.1269454993769,59.0921363793662,59.098609919398,59.0683326693666,59.0678077274881,59.0591659493731,59.0408325194264,59.0161132794218,58.9971961994622,58.950927729477,58.9815826395124,58.9506835895355]}],[{"lng":[-70.3394470199719,-70.314163209971,-70.3936080899736,-70.4302749599749,-70.4177780199746,-70.3394470199719],"lat":[47.4272232099092,47.4249992399047,47.3688888499182,47.3699989299238,47.4094428999219,47.4272232099092]}],[{"lng":[-69.3611602801947,-69.3467865002009,-69.3800201401857,-69.3611602801947],"lat":[59.0394592295706,59.0095176695612,58.9925117495815,59.0394592295706]}],[{"lng":[-69.4181137101695,-69.3907470701808,-69.4049987801744,-69.4458236701586,-69.422225950168,-69.4181137101695],"lat":[58.9791679396039,58.9731101995877,58.9444427495958,58.9723663296197,58.9883346596064,58.9791679396039]}],[{"lng":[-70.4225006099739,-70.5124969499768,-70.5338897699774,-70.5752792399787,-70.5136108399769,-70.4225006099739],"lat":[47.167778019923,47.1047210699356,47.0555572499384,47.0402793899434,47.1213874799358,47.167778019923]}],[{"lng":[-69.5803756701206,-69.6044998201136,-69.5805130001207,-69.5803756701206],"lat":[59.7005271896973,59.7012939497083,59.7149200396976,59.7005271896973]}],[{"lng":[-70.61833190998,-70.6383361799806,-70.6091690099798,-70.61833190998],"lat":[47.0400009199482,47.0272216799503,47.0491676299472,47.0400009199482]}],[{"lng":[-69.6633300800998,-69.6383056601063,-69.645706180104,-69.6916961700926,-69.6633300800998],"lat":[59.9575042697363,59.9469451897258,59.9091606097285,59.9174575797474,59.9575042697363]}],[{"lng":[-70.7352752699834,-70.7602767899841,-70.7108306899828,-70.7352752699834],"lat":[47.0111122099596,47.0013885499618,47.0244445799574,47.0111122099596]}],[{"lng":[-71.0952072099914,-71.1447219799922,-71.128334049992,-70.9405517599885,-70.8499984699864,-70.7979431199851,-70.8874969499872,-71.0072250399897,-71.0952072099914],"lat":[46.8485259999826,46.8511123699846,46.8816680899839,46.9961128199746,47.0246658299687,47.0248870799648,46.9197235099713,46.8574981699784,46.8485259999826]}],[{"lng":[-69.9104156500562,-69.9224014300545,-69.8979187000583,-69.9104156500562],"lat":[60.9500007598303,60.9682311998338,60.972915649827,60.9500007598303]}],[{"lng":[-69.9479141200511,-69.9046859700573,-69.9494781500507,-69.9401016200519,-69.9791641200464,-70.0244827300411,-69.9479141200511],"lat":[61.0395851098414,61.0005187998291,60.9828147898413,60.9651069598387,60.9312515298487,60.9953155498604,61.0395851098414]}],[{"lng":[-70.0374984700393,-70.0270843500404,-70.0500030500379,-70.0374984700393],"lat":[60.8979148898629,60.8770828198603,60.9104156498659,60.8979148898629]}],[{"lng":[-71.5091781599998,-71.4921874999998,-71.5703124999998,-71.5786514299998,-71.5091781599998],"lat":[61.3329200699951,61.3005218499949,61.2984352099959,61.324478149996,61.3329200699951]}],[{"lng":[-71.5791702299998,-71.5609359699998,-71.5994796799998,-71.5791702299998],"lat":[61.6708335899961,61.6609344499959,61.6588516199963,61.6708335899961]}],[{"lng":[-71.6171874999998,-71.5984344499998,-71.6249999999998,-71.6171874999998],"lat":[61.3140602099965,61.3088569599963,61.3020820599966,61.3140602099965]}],[{"lng":[-71.7104186999998,-71.6817703199998,-71.7005157499998,-71.7494811999998,-71.7104186999998],"lat":[61.5291671799974,61.5203094499972,61.5067710899973,61.5130195599978,61.5291671799974]}],[{"lng":[-72.0916671799999,-71.9609374999999,-72.0791702299999,-72.1286468499999,-72.0916671799999],"lat":[61.8916664099994,61.858856199999,61.8604164099994,61.8755187999995,61.8916664099994]}],[{"lng":[-72.1838531499999,-72.1526031499999,-72.1973953199999,-72.1838531499999],"lat":[61.9953155499996,61.9776039099995,61.9838561999996,61.9953155499996]}],[{"lng":[-72.27500153,-72.30781555,-72.27708435,-72.27500153],"lat":[61.9958343499997,62.0015602099998,62.0083351099997,61.9958343499997]}],[{"lng":[-72.4375,-72.32968903,-72.41666412,-72.46198273,-72.4375],"lat":[62.0437507599999,62.0130195599998,62.0125007599999,62.0276069599999,62.0437507599999]}],[{"lng":[-72.42760468,-72.39427185,-72.43489838,-72.42760468],"lat":[62.0703124999999,62.0567741399998,62.0609397899999,62.0703124999999]}],[{"lng":[-73.00150299,-72.97644806,-72.98088837,-73.0349884,-73.088974,-73.14318848,-73.13757324,-73.08638,-73.00150299],"lat":[46.15731049,46.14860535,46.13453674,46.12413025,46.07950211,46.06280518,46.10302734,46.13592529,46.15731049]}],[{"lng":[-72.8354187,-72.79426575,-72.87239838,-72.8354187],"lat":[62.23958206,62.22135162,62.2338562,62.23958206]}],[{"lng":[-73.09791565,-73.06718445,-73.125,-73.13906097,-73.09791565],"lat":[62.27083206,62.25885391,62.25,62.26197815,62.27083206]}],[{"lng":[-74.00468445,-74.15208435,-74.30833435,-74.37916565,-74.54582977,-74.60364532,-74.47916412,-74.4151001,-74.33125305,-74.1791687,-73.98750305,-73.95885468,-73.98281097,-73.92343903,-74.00468445],"lat":[62.59635162,62.59583282,62.61875153,62.64583206,62.6645851099999,62.6973991399999,62.7229156499999,62.71197891,62.66249847,62.68125153,62.66458511,62.63489914,62.62968445,62.61302185,62.59635162]}],[{"lng":[-75.5453109700001,-75.5458297700001,-75.5765609700001,-75.5453109700001],"lat":[62.2921905499945,62.2750015299945,62.281768799994,62.2921905499945]}],[{"lng":[-76.650001529981,-76.6598968499804,-76.6005172699837,-76.6682281499799,-76.66822814998,-76.5979156499839,-76.5421829199865,-76.5890655499843,-76.5895843499843,-76.6578140299806,-76.6276016199824,-76.7187499999768,-77.0348968499433,-77.0276031499443,-77.0828170799355,-77.0630187999388,-77.1473998999237,-77.1380157499254,-77.1932296799142,-77.2192687999082,-77.403648379856,-77.3958358798584,-77.2880172698908,-77.4083328198542,-77.4328155498454,-77.3994827298569,-77.3067703198853,-77.3104171798843,-77.2671890298959,-77.2979202298879,-77.1708297699182,-77.0833358799348,-77.1078109699304,-77.0312499999431,-76.9401016199555,-76.9958343499483,-76.9083328199593,-76.8026046799698,-76.752601619974,-76.7682342499728,-76.66510009998,-76.7057342499774,-76.7015609699777,-76.650001529981],"lat":[63.5250015299324,63.510940549931,63.5046844499384,63.4807281499298,63.4588546799297,63.4625015299385,63.4463539099448,63.4369811999395,63.4208335899394,63.4223937999309,63.3963546799347,63.3624992399223,63.4213561998643,63.4369811998661,63.4484405498531,63.451564789858,63.4838561998369,63.4932289098395,63.5026016198245,63.5390624998174,63.5859374997568,63.61249923976,63.6161460897972,63.6166648897555,63.6369781497465,63.6661491397595,63.685939789792,63.697917939791,63.6828155498046,63.6729164097947,63.6833343498327,63.6520843498549,63.674480439849,63.6749992398672,63.6453094498862,63.6354179398747,63.6354179398923,63.6119804399107,63.5848960899184,63.5755195599159,63.5557289099305,63.551559449925,63.5234374999254,63.5250015299324]}],[{"lng":[-76.0400009200019,-76.0244445800019,-76.0755538900019,-76.0400009200019],"lat":[56.1652793899751,56.154167179976,56.1758346599729,56.1652793899751]}],[{"lng":[-76.1215896600018,-76.0855712900019,-76.0800170900019,-76.1215896600018],"lat":[56.2354621899699,56.2282867399723,56.2125244099726,56.2354621899699]}],[{"lng":[-76.1488876300018,-76.1361084000018,-76.1688919100017,-76.1488876300018],"lat":[56.247222899968,56.2395019499689,56.2472228999665,56.247222899968]}],[{"lng":[-76.3188858000012,-76.3816680900007,-76.347778320001,-76.1622085600018,-76.1710815400018,-76.3188858000012],"lat":[56.1188888499534,56.1405563399468,56.1636123699505,56.1425514199669,56.1317138699662,56.1188888499534]}],[{"lng":[-76.2811279300012,-76.1843872100017,-76.2450027500015,-76.3461074800008,-76.2811279300012],"lat":[56.2739257799571,56.2469444299653,56.2436103799604,56.2708320599508,56.2739257799571]}],[{"lng":[-76.312416080001,-76.2811279300012,-76.3377761800009,-76.312416080001],"lat":[56.2861595199542,56.2824706999572,56.2802772499516,56.2861595199542]}],[{"lng":[-76.3649978600005,-76.3438797000007,-76.3944473300002,-76.3649978600005],"lat":[56.3583335899489,56.3544998199511,56.3563880899457,56.3583335899489]}],[{"lng":[-76.7061309799913,-76.7390136699898,-76.7089538599909,-76.6705474899925,-76.6899032599918,-76.6423797599935,-76.6780624399924,-76.6466903699935,-76.6466140699936,-76.692413329992,-76.7061309799913],"lat":[57.1118240399014,57.1727294898953,57.2013435399011,57.1694335899079,57.1565284699045,57.1380538899126,57.1075363199064,57.0899772599118,57.0661811799118,57.0654640199038,57.1118240399014]}],[{"lng":[-76.6355895999953,-76.6600036599946,-76.6288909899952,-76.6394042999947,-76.6082763699957,-76.6294479399951,-76.6133346599958,-76.6355895999953],"lat":[56.5586547899126,56.5394439699084,56.6630554199139,56.7175292999123,56.7025146499173,56.6780548099138,56.5794448899163,56.5586547899126]}],[{"lng":[-76.6500015299943,-76.6166686999953,-76.6205520599953,-76.661666869994,-76.6500015299943],"lat":[56.7619438199106,56.7472228999161,56.7161102299154,56.7338905299085,56.7619438199106]}],[{"lng":[-76.7054824799915,-76.6450958299938,-76.6994476299919,-76.7054824799915],"lat":[57.0584182699014,57.0210380599119,56.9908332799023,57.0584182699014]}],[{"lng":[-76.648887629994,-76.6405563399944,-76.6755523699932,-76.648887629994],"lat":[56.879444119911,56.8527793899124,56.8461112999063,56.879444119911]}],[{"lng":[-77.4005203198563,-77.3796844498633,-77.4208297698495,-77.4432296798412,-77.4005203198563],"lat":[63.6973991397596,63.6901016197671,63.6645851097515,63.6869811997433,63.6973991397596]}],[{"lng":[-76.6594238299934,-76.6622238199934,-76.6939086899923,-76.6938857999922,-76.6594238299934],"lat":[56.9888915999094,56.9586791999089,56.9522705099033,56.9868774399033,56.9888915999094]}],[{"lng":[-76.6552047699949,-76.618896479996,-76.6399993899955,-76.6766967799944,-76.686668399994,-76.6552047699949],"lat":[56.4756660499091,56.4525146499152,56.4261093099116,56.4295043899053,56.4519462599035,56.4756660499091]}],[{"lng":[-76.6455459599951,-76.6633300799946,-76.6660690299944,-76.6455459599951],"lat":[56.5222816499108,56.5099983199078,56.5272445699073,56.5222816499108]}],[{"lng":[-76.7559661899887,-76.7205810499903,-76.7219848599904,-76.7834091199873,-76.7559661899887],"lat":[57.2719917298923,57.248622889899,57.2058715798987,57.2676048298867,57.2719917298923]}],[{"lng":[-77.4458312998404,-77.4276046798471,-77.4687499998317,-77.4458312998404],"lat":[63.670833589742,63.6630210897489,63.6562499997327,63.670833589742]}],[{"lng":[-76.6972198499922,-76.703308109992,-76.6950073199923,-76.6972198499922],"lat":[56.9180564899026,56.9088745099014,56.931518549903,56.9180564899026]}],[{"lng":[-76.7300949099898,-76.7659072899882,-76.8396072399836,-76.7446289099889,-76.7300949099898],"lat":[57.2849121098974,57.2802352898903,57.4312972998751,57.3683853098948,57.2849121098974]}],[{"lng":[-76.6650009199949,-76.6855544999943,-76.6652831999949,-76.6650009199949],"lat":[56.3713874799072,56.3586120599035,56.3827438399072,56.3713874799072]}],[{"lng":[-77.4458312998406,-77.4682311998321,-77.4541702298374,-77.4458312998406],"lat":[63.6291656497413,63.6296844497324,63.6458320597383,63.6291656497413]}],[{"lng":[-76.7343673699928,-76.6738891599947,-76.668891909995,-76.7460937499924,-76.7595825199918,-76.7343673699928],"lat":[56.2944755598942,56.3508911099056,56.2988891599064,56.2739257798918,56.2819213898891,56.2944755598942]}],[{"lng":[-76.7688217199919,-76.7583312999922,-76.7055587799943,-76.7866821299913,-76.7688217199919],"lat":[56.1544418298869,56.1819457998892,56.1322212198994,56.0996704098831,56.1544418298869]}],[{"lng":[-77.512496949817,-77.5619811997953,-77.5067672698197,-77.5536498997992,-77.5338516198082,-77.5854186997845,-77.639060969757,-77.6046829197751,-77.6708297697394,-77.8666686996036,-77.9395828195386,-77.9421844495358,-77.9958343494819,-78.0255203194491,-78.0911483793714,-78.088020319375,-78.3494796789591,-78.3526000989523,-78.4723968486902,-78.5286483785461,-78.1791686992441,-78.0479202294174,-78.0104141194596,-77.9145812995562,-77.895835879573,-77.8437499996173,-77.8145828196401,-77.7734374996706,-77.8807296795868,-77.74583434969,-77.693748469723,-77.6380157497551,-77.6744842497347,-77.6213531497643,-77.6161498997674,-77.5234374998118,-77.5474014298015,-77.5124969498166,-77.4755172698317,-77.5223998998125,-77.4755172698319,-77.512496949817],"lat":[63.2333335897071,63.1984405496842,63.1921844497088,63.1911430396879,63.1838569596967,63.1624984696724,63.1619758596458,63.1463508596627,63.147914889629,63.0895843495103,63.0895843494595,63.1098937994582,63.110416409418,63.1411437993958,63.1526069593421,63.1661491393453,63.3026008590965,63.320312499094,63.3630218489553,63.4307289088876,63.4833335892778,63.4583320593895,63.4708328194194,63.4541664094887,63.4645843495018,63.4520835895357,63.4604148895543,63.4359397895784,63.4213561995107,63.4020843495936,63.4208335896229,63.4161491396518,63.3984641996326,63.3911437996595,63.3442687996611,63.3036460897035,63.2651061996921,63.2750015297078,63.2661437997234,63.2619781497033,63.2473983797231,63.2333335897071]}],[{"lng":[-76.733886719993,-76.7177734399936,-76.7666015599918,-76.7466659499926,-76.7783355699912,-76.733886719993],"lat":[56.2366676298941,56.2122878998972,56.1939086898875,56.2099990798916,56.2377777098851,56.2366676298941]}],[{"lng":[-76.9078063999783,-76.8656005899815,-76.8779449499809,-76.9078063999783],"lat":[57.5834197998596,57.5487060498696,57.4987640398665,57.5834197998596]}],[{"lng":[-77.0438919099649,-77.0138854999681,-77.0150146499682,-77.0438919099649],"lat":[57.7900009198235,57.7788886998323,57.7496948198318,57.7900009198235]}],[{"lng":[-77.7562484696928,-77.7375030497052,-77.6374969497635,-77.7499999996976,-77.7916641196692,-77.8286514296412,-77.7562484696928],"lat":[62.5895843495676,62.5791664095785,62.5833320596347,62.53541564957,62.5354156495442,62.5869789095216,62.5895843495676]}],[{"lng":[-77.4255523698761,-77.3861083998879,-77.4619750998645,-77.4255523698761],"lat":[59.8764038096967,59.843612669714,59.8765258796794,59.8764038096967]}],[{"lng":[-77.4469146698695,-77.4205551098778,-77.4708175698617,-77.4469146698695],"lat":[59.8622207596865,59.8466682396986,59.8619193996749,59.8622207596865]}],[{"lng":[-77.9604186995272,-77.8713531496073,-77.8484344496264,-77.9145812995709,-78.0500030494333,-78.0583343494235,-78.0624999994189,-78.1078109693637,-78.1119842493579,-78.0666656494136,-78.0604171794206,-77.9604186995272],"lat":[62.591667179428,62.5869789094928,62.5484352095074,62.5374984694606,62.5562515293547,62.5687484693481,62.5562515293441,62.5630187993045,62.5807304393014,62.5750007593412,62.5916671793472,62.591667179428]}],[{"lng":[-77.5022201498517,-77.4927749598552,-77.5213165298447,-77.5022201498517],"lat":[59.7719459496581,59.7569427496627,59.7788200396483,59.7719459496581]}],[{"lng":[-77.8526000996366,-77.806770319671,-77.8249969496581,-77.8807296796144,-77.8526000996366],"lat":[61.6765594494812,61.6515655495124,61.6333351094995,61.6692695594607,61.6765594494812]}],[{"lng":[-77.1843109099615,-77.1391982999669,-77.2378845199545,-77.2258071899559,-77.1843109099615],"lat":[55.7708625797669,55.7672347997835,55.7335281397457,55.7671241797507,55.7708625797669]}],[{"lng":[-77.6479186997854,-77.6291656497946,-77.7140655497508,-77.6479186997854],"lat":[60.2083320595839,60.1916656495948,60.1984405495425,60.2083320595839]}],[{"lng":[-77.727081299743,-77.652603149783,-77.6729202297728,-77.727081299743],"lat":[60.2333335895346,60.2151069595812,60.2083320595687,60.2333335895346]}],[{"lng":[-77.8963546796015,-77.872917179621,-77.9286498995736,-77.8963546796015],"lat":[61.6744804394492,61.656249999466,61.6692695594245,61.6744804394492]}],[{"lng":[-77.217971799958,-77.1857910199622,-77.2553100599529,-77.2517089799536,-77.3803100599318,-77.217971799958],"lat":[55.6515159597533,55.6533203097659,55.619079589738,55.6002807597394,55.5521240196809,55.6515159597533]}],[{"lng":[-78.0520858794335,-78.0171890294723,-78.0583343494266,-78.0520858794335],"lat":[62.4354171793486,62.4296874993776,62.4249992393429,62.4354171793486]}],[{"lng":[-77.5916519198366,-77.5106811498661,-77.5250015298614,-77.5916519198366],"lat":[58.2239456195883,58.2086791996345,58.2000007596265,58.2239456195883]}],[{"lng":[-77.7321929897476,-77.7105712897597,-77.72833251975,-77.7630004897296,-77.7321929897476],"lat":[59.7327499395217,59.724906919536,59.7177772495241,59.7338867195006,59.7327499395217]}],[{"lng":[-77.5677795398463,-77.6865310697952,-77.5311126698596,-77.5677795398463],"lat":[58.1773071296019,58.2301864595279,58.1727790796228,58.1773071296019]}],[{"lng":[-77.5561141998509,-77.5380401598575,-77.5857772798396,-77.5561141998509],"lat":[58.1602783196085,58.1440238996186,58.1656761195911,58.1602783196085]}],[{"lng":[-78.4583358787175,-78.4494781487389,-78.4255218487931,-78.4088516188314,-78.5187530485714,-78.5432281485065,-78.5223998985601,-78.4583358787175],"lat":[63.5166664089824,63.4963569589918,63.503646849021,63.4567718490378,63.4416656489013,63.4484405488695,63.4828147888993,63.5166664089824]}],[{"lng":[-77.6360092198642,-77.3814315799316,-77.4741058299117,-77.6360092198642],"lat":[55.4301452595317,55.5512084996804,55.4798545796315,55.4301452595317]}],[{"lng":[-77.4999389598918,-77.4749145498987,-77.4988861098929,-77.5232696498859,-77.544441219879,-77.4949951198935,-77.4999389598918],"lat":[56.4342384296236,56.4439620996376,56.3786125196238,56.3725738496098,56.3961105295976,56.4166870096263,56.4342384296236]}],[{"lng":[-77.8041915897309,-77.6963653597901,-77.6908264197931,-77.7447128297655,-77.8158264197239,-77.8041915897309],"lat":[58.2742233294436,58.2467193595215,58.229442599525,58.2374992394874,58.2666931194344,58.2742233294436]}],[{"lng":[-78.1151046793994,-78.0734329194484,-78.1562499993492,-78.1828155493134,-78.1151046793994],"lat":[60.7890624992325,60.7671852092722,60.7458343491892,60.7682304391621,60.7890624992325]}],[{"lng":[-77.9816894496017,-78.0073699995793,-77.8911132796726,-77.7711181597503,-77.8340530397122,-77.9816894496017],"lat":[58.3239135692928,58.3261833192683,58.3194427493739,58.2739257794686,58.2781715394202,58.3239135692928]}],[{"lng":[-78.616149898452,-78.6640624983137,-78.6765670782744,-78.621353148435,-78.6020812984867,-78.4749984687987,-78.4479141188584,-78.4515609688496,-78.4187469489175,-78.2401046792298,-78.2817687991664,-78.4015655489541,-78.318748469106,-78.616149898452],"lat":[60.7088546785767,60.7088546784957,60.7307281484753,60.7484397885707,60.775001528604,60.7958335887997,60.7791671788372,60.7994804388332,60.8083343488789,60.8244781491013,60.7859344490516,60.7567710888995,60.7583351090058,60.7088546785767]}],[{"lng":[-77.7389907798037,-77.7362060498056,-77.7584838897946,-77.7389907798037],"lat":[56.3775901794668,56.3508911094687,56.3651199294519,56.3775901794668]}],[{"lng":[-78.0614852895274,-77.9888915995952,-78.035675049553,-78.083885189505,-78.0614852895274],"lat":[58.3648719792152,58.3353271492862,58.3375701892407,58.3619461091918,58.3648719792152]}],[{"lng":[-78.1488876294072,-78.1422195394155,-78.1688842793831,-78.1488876294072],"lat":[59.1658325191443,59.1566657991513,59.1588745091219,59.1658325191443]}],[{"lng":[-78.1379757639055,-78.2044067393653,-78.1099471994763,-78.0671997095216,-78.1379757639055],"lat":[58.3799367873224,58.4051437390578,58.4004211391651,58.3690795892094,58.3799367873224]}],[{"lng":[-77.965553279671,-77.8766326897302,-77.8861083997246,-77.8344116197554,-77.9127807597081,-77.9310913096954,-77.971710209667,-77.965553279671],"lat":[56.2999992392712,56.327999109354,56.3163871793454,56.3236122093903,56.288333889321,56.3038940393042,56.2836303692649,56.2999992392712]}],[{"lng":[-78.3043670692,-78.3006744392066,-78.3443908691368,-78.3383178691457,-78.3043670692],"lat":[59.0080947889531,58.9875717189571,58.9780845588988,59.0053100589081,59.0080947889531]}],[{"lng":[-78.035499569616,-78.0062218842064,-78.0723495495849,-78.076110839581,-78.035499569616],"lat":[56.3074645992004,56.3028828759079,56.2835235591604,56.2978515591565,56.3074645992004]}],[{"lng":[-78.0061012219811,-77.9950485196487,-78.005493159641,-78.0061012219811],"lat":[56.3028639764838,56.3011321992419,56.2886962892312,56.3028639764838]}],[{"lng":[-78.0560455295902,-78.0460510295994,-78.0693664595785,-78.0560455295902],"lat":[56.5487594591827,56.5376472491931,56.5432128891682,56.5487594591827]}],[{"lng":[-78.4683151189108,-78.4634017889187,-78.454711908937,-78.4337234489779,-78.4426269489612,-78.4164962790105,-78.4069213890288,-78.4105834990213,-78.3777542090808,-78.4029159490373,-78.4683151189108],"lat":[58.9174423187173,58.9556541387265,58.9410476687391,58.9474296587709,58.9359054587571,58.9515304587964,58.9413185088099,58.9553222688051,58.9534797688519,58.9197158788148,58.9174423187173]}],[{"lng":[-78.3662261991224,-78.3168945292034,-78.395301819072,-78.3662261991224],"lat":[58.5022926288509,58.4822997989179,58.5012474088093,58.5022926288509]}],[{"lng":[-78.1126785295366,-78.0910186795578,-78.1265716595231,-78.1126785295366],"lat":[56.5737419091206,56.5637435891448,56.5665206891045,56.5737419091206]}],[{"lng":[-78.3098525992731,-78.2498779293562,-78.2949218692952,-78.3099060092749,-78.371665949181,-78.3599853491983,-78.3098525992731],"lat":[57.2097129788878,57.1912841789675,57.1933669989078,57.1717605588867,57.1725006087987,57.1951789888164,57.2097129788878]}],[{"lng":[-78.2971801792889,-78.314941409264,-78.3383331292289,-78.2971801792889],"lat":[57.2597045889065,57.2456054688818,57.2511100788491,57.2597045889065]}],[{"lng":[-78.4040832491192,-78.3495330792076,-78.3046874992755,-78.425415039085,-78.4807891789819,-78.4040832491192],"lat":[57.3242111187551,57.3328857388356,57.3154907188978,57.2750854487208,57.2861251786331,57.3242111187551]}],[{"lng":[-78.4902343688831,-78.5013503988605,-78.5308227487958,-78.4902343688831],"lat":[58.6064605686695,58.5919456486511,58.5996704086034,58.6064605686695]}],[{"lng":[-78.5991821286317,-78.5308837887935,-78.4942932088739,-78.5775909386874,-78.6164855985896,-78.5991821286317],"lat":[58.6378822284885,58.6350707986049,58.6179122886635,58.5975723285243,58.6142387384565,58.6378822284885]}],[{"lng":[-78.5525436387341,-78.5346145587763,-78.5755844086808,-78.5525436387341],"lat":[58.7925071685763,58.7769317586054,58.7699623085361,58.7925071685763]}],[{"lng":[-78.6255187985518,-78.5844116186565,-78.5883178686456,-78.5695648186928,-78.5467224087458,-78.5733032186849,-78.5836868286589,-78.6134490985846,-78.6255187985518],"lat":[58.8159484884506,58.8117065385229,58.8317260685172,58.8093795785483,58.8208198485873,58.791667938541,58.8027076685237,58.7962608284712,58.8159484884506]}],[{"lng":[-78.6000976586199,-78.5856704686568,-78.6181640585751,-78.6000976586199],"lat":[58.7763061484938,58.7611122085182,58.7583427384609,58.7763061484938]}],[{"lng":[-78.6906585683865,-78.6499023385006,-78.64690398851,-78.5730133087,-78.6906585683865],"lat":[58.6277160583193,58.6265258783962,58.6091041584009,58.574104308531,58.6277160583193]}],[{"lng":[-78.4495696990354,-78.4111328091049,-78.5222244288946,-78.4495696990354],"lat":[57.3778037986865,57.3629531887456,57.3502540585664,57.3778037986865]}],[{"lng":[-78.6885452283798,-78.6680145284383,-78.7140884383029,-78.668014528437,-78.6235961885588,-78.6588592484655,-78.6510009784861,-78.7063445983287,-78.6885452283798],"lat":[58.7899665783324,58.7910461383713,58.807369228284,58.8079910283722,58.7882690384527,58.7678565983871,58.7813262884024,58.7754936182973,58.7899665783324]}],[{"lng":[-78.6721801784146,-78.6469116184856,-78.6917037983585,-78.6721801784146],"lat":[58.9489135683723,58.9406738284185,58.9450035083351,58.9489135683723]}],[{"lng":[-78.4082412691473,-78.3360137892609,-78.4396972690938,-78.4082412691473],"lat":[56.70513152873,56.6937408388371,56.7017211886807,56.70513152873]}],[{"lng":[-78.6862106283762,-78.6701202384226,-78.693481448356,-78.6862106283762],"lat":[58.9240951483444,58.9199371283745,58.9102134683298,58.9240951483444]}],[{"lng":[-78.6305236785509,-78.6443786585153,-78.7011108383542,-78.715911868307,-78.6915893583788,-78.693840028374,-78.6855087283972,-78.6305236785509],"lat":[58.6455650284328,58.6282157884065,58.6505126983004,58.6895141582736,58.690307618321,58.6688957183155,58.679176328332,58.6455650284328]}],[{"lng":[-80.2583312796226,-80.2723998791846,-80.2036514109413,-80.1244811828552,-80.0749969339783,-80.089065533669,-80.0588531343372,-80.0661468341657,-79.985939015814,-79.9578170663419,-79.8541641081974,-79.7583312897181,-79.7994842390894,-79.7791671694077,-79.739585869995,-79.7229156402397,-79.604164111796,-79.4609374933933,-79.4473953135334,-79.4026031439631,-79.4369811936378,-79.3588485643615,-79.3578109643771,-79.2921829149251,-79.3286514246269,-79.3124999947629,-79.2713546750895,-79.260932915177,-79.3015670748615,-79.2583312952043,-79.2963485649183,-79.3223953147068,-79.3067703148373,-79.3369827245937,-79.3005218448998,-79.4578170735111,-79.425521843832,-79.5390624926919,-79.5338516127624,-79.5942687920884,-79.672401421119,-79.6755218410928,-79.7015609607449,-79.6671829112055,-79.7057342406908,-79.6859359609623,-79.7255172604301,-79.8062515192473,-79.8494796685568,-79.9036483676196,-79.8963546677336,-79.9328155370854,-79.9338531370506,-79.9619827165258,-79.9380187869709,-79.9890670659951,-79.9525985566967,-79.9994811857799,-79.9651031364496,-79.9849014160614,-79.9630203064829,-80.0500030347193,-80.0619811844672,-80.058334334539,-80.0828170640075,-80.0374984549999,-80.0854186839572,-80.1848983616164,-80.2119827109182,-80.1921844314104,-80.2494811799047,-80.2583312796226],"lat":[61.9145851030653,61.9973983730116,62.1536483734702,62.224475853931,62.2354164041945,62.2255210841184,62.2234344442736,62.2515602042451,62.3088569546611,62.3619804348104,62.3916664052876,62.3916664056896,62.3869781455193,62.3770828156011,62.3895835857644,62.3729171758266,62.4145851062822,62.3828124967617,62.3567695568003,62.3536491369393,62.344268796831,62.3307304370671,62.292190547064,62.2869758572521,62.2890624971487,62.2750015271929,62.2807311973091,62.2296905473303,62.2130241372146,62.1791648873302,62.1171874972151,62.1182289071405,62.1005210871829,62.0661430370899,62.0234374971893,61.9453124966948,61.9026069567919,61.8078155463944,61.7484397864017,61.6984405461744,61.691146845875,61.6380195558516,61.6401061957481,61.6234397858814,61.6307258557292,61.613021845805,61.5880241356391,61.5687484652916,61.5734405451001,61.6234397848628,61.6598930349065,61.6546897847325,61.6953124947385,61.6984405446031,61.7036437947207,61.7192687944752,61.7130241346529,61.7421874944297,61.7401008545994,61.7557258545062,61.7557258546139,61.7958335841877,61.7880210841228,61.8020820541462,61.8005218440168,61.7562484642405,61.7854156439982,61.7859382534492,61.8047294532995,61.8286437934221,61.8442687930926,61.9145851030653]}],[{"lng":[-78.517349238892,-78.5185852088908,-78.546997068829,-78.517349238892],"lat":[57.5444984385819,57.523403168579,57.5401000985311,57.5444984385819]}],[{"lng":[-78.5888290387375,-78.6100158686883,-78.6457748385993,-78.5888290387375],"lat":[57.4973831184553,57.482593538416,57.483943938349,57.4973831184553]}],[{"lng":[-78.9334411575101,-78.927864067531,-78.8805541977299,-78.9407958974804,-78.9334411575101],"lat":[58.848892207811,58.8727950978261,58.8044433579314,58.8350830077923,58.848892207811]}],[{"lng":[-78.7100830084233,-78.7382202083432,-78.7243881183811,-78.7100830084233],"lat":[57.5164794882239,57.4996948181653,57.5294418281954,57.5164794882239]}],[{"lng":[-78.7350845283925,-78.7159957884469,-78.7537994383382,-78.7350845283925],"lat":[57.0641593881514,57.0632247881911,57.0576591481115,57.0641593881514]}],[{"lng":[-78.7129898085129,-78.6652831986373,-78.6519775386722,-78.6492156986911,-78.681114198612,-78.6705932586398,-78.6941833485798,-78.655914308677,-78.658645628673,-78.6860809286043,-78.6691894486507,-78.7219619785138,-78.7553253184234,-78.7777938783561,-78.8171234082405,-78.9086608879346,-78.9208526578878,-78.8831405580181,-78.9153289779059,-78.8969955379664,-78.7860107383189,-78.8503265381175,-78.8059081982566,-78.8336639381664,-78.8112716682357,-78.8375396681509,-78.7672119083618,-78.7129898085129],"lat":[56.4356841981715,56.4337158182688,56.4122924782944,56.2692336982949,56.268333438231,56.2539062482517,56.2521629282037,56.2442474382807,56.2128181482742,56.2126617382189,56.1708984382517,56.1703529381428,56.1470184280702,56.1672515880214,56.1289634679306,56.1078529377087,56.1263313276787,56.1648101777747,56.1436843876935,56.1884078977415,56.2883911080078,56.2678527778585,56.3015136679634,56.3195190379,56.3403129579527,56.345283507892,56.4155273380547,56.4356841981715]}],[{"lng":[-78.7997970581648,-78.7890319782007,-78.8466186480093,-78.7997970581648],"lat":[57.3681716880265,57.3553390480495,57.3508911079197,57.3681716880265]}],[{"lng":[-78.7579956083496,-78.7351760884161,-78.7390899684063,-78.7680053683205,-78.7579956083496],"lat":[56.8120727480915,56.8098792981401,56.7943305981312,56.8035278280694,56.8120727480915]}],[{"lng":[-78.7838745082802,-78.7262420684497,-78.8061141982112,-78.7838745082802],"lat":[56.7258911080311,56.7211265581552,56.7213897679811,56.7258911080311]}],[{"lng":[-78.8307418781362,-78.745483398399,-78.7349395784308,-78.8166122381845,-78.856056208052,-78.8307418781362],"lat":[56.6783752379228,56.6788940381127,56.6622047381344,56.6572036679543,56.6713714578631,56.6783752379228]}],[{"lng":[-78.7820968582996,-78.7709960883344,-78.8120956382072,-78.7820968582996],"lat":[56.5891685480291,56.575317378053,56.5797233579612,56.5891685480291]}],[{"lng":[-78.7704772883812,-78.7622222884063,-78.816665648245,-78.7704772883812],"lat":[56.1322746280363,56.1208343480541,56.1013870179305,56.1322746280363]}],[{"lng":[-78.9810790976735,-79.0531234673914,-79.1186141970989,-79.0023803675794,-78.946105957798,-78.9397041783309,-78.9011764479671,-78.8289184582045,-78.8214111282313,-78.7794418283573,-78.7896270783282,-78.9371643078371,-78.9386636421926,-78.9690551777155,-78.9334106378535,-78.9877929676432,-79.0280837974824,-78.9738769476996,-78.9038848879628,-78.9810790976735],"lat":[56.0000152575153,55.8546676573079,55.7839012071104,56.0504760674599,56.0900878876121,56.0739645799997,56.0635833677256,56.1173095679027,56.0902709979191,56.1100006080155,56.0981407179922,56.0505371076335,56.0563289978605,56.0411415075494,56.0330810476424,56.0230712874983,55.9730567873848,56.0184936475355,56.0210876477169,56.0000152575153]}],[{"lng":[-78.7512741084721,-78.7661132784313,-78.7492599484749,-78.7512741084721],"lat":[55.781311038066,55.7727394080329,55.8081245380713,55.781311038066]}],[{"lng":[-79.9027215272929,-79.8873901303281,-79.9091186400174,-79.9275207397662,-79.9319686796803,-79.9474792394544,-79.9513397093909,-79.9730834890591,-79.9675292891399,-79.9871597188348,-79.9779968189721,-80.01879881832,-80.0122070184116,-79.9661102191315,-79.9799880889223,-79.8842010403207,-79.8331909110136,-79.864685050588,-79.7594146618912,-79.5363769441698,-79.4548721248144,-79.4748687646477,-79.4531860347985,-79.4115371651566,-79.4725952046958,-79.5187606743425,-79.5952758736501,-79.6004028236151,-79.6415328932178,-79.5136108343964,-79.4315338050666,-79.3970947253206,-79.2975845259484,-79.1203155469706,-79.0332031173927,-78.9251098578332,-78.9323883078167,-78.9049987779184,-78.9412841777846,-78.953659057742,-78.9266433678416,-78.9151534978861,-78.8823852480027,-78.9262771578478,-78.9584960877252,-78.9590530377218,-79.2130432065918,-79.1185378970521,-78.9893951376063,-78.9545440677307,-79.0067977875175,-78.9700012176668,-78.9933319075707,-78.9390869077851,-79.0050048775211,-78.9907760575828,-79.055580137304,-79.095825197137,-79.0986938471277,-79.0885897819757,-79.0632171572922,-79.0547790473328,-79.1043548571042,-79.208595276602,-79.2830352761805,-79.1278991669944,-79.1315002369563,-79.1737289367382,-79.2256774864583,-79.3558120656769,-79.4897003146919,-79.5451278642162,-79.6139831435549,-79.7314605623478,-79.7830352717472,-79.6436767532537,-79.6363654625238,-79.466125484819,-79.4756545947332,-79.5162582343878,-79.8131103413234,-79.8598403807392,-79.8796005104637,-79.9030380101435,-79.9100952000259,-79.9027215272929],"lat":[55.8576792152289,55.8948974538479,55.8972778237241,55.8622207536133,55.9014244035919,55.8877792334989,55.8970947234771,55.8890991133458,55.9000434833807,55.8924713032608,55.9071922233184,55.8925743030655,55.9232788031105,55.9575004533962,55.9411697333105,56.0036354038784,56.0061035141609,56.0114746039885,56.1254997245627,56.2672958356226,56.4482612559756,56.4571495058958,56.5596923759919,56.4424247661452,56.3505248958959,56.2118797256927,56.1717414853554,56.1262817353282,56.0849266051353,56.1809081957118,56.1935386660473,56.2207603461832,56.4972419665685,56.4861564571445,56.4221496573937,56.4291992176821,56.3439216576594,56.3430557277286,56.3300781276359,56.2901458676018,56.3188438376729,56.3045234677013,56.3136444077831,56.280864717672,56.2744750975884,56.2839202875874,55.8561134268144,56.0576362571251,56.2369537375043,56.3489990176024,56.3510971074629,56.3686103775628,56.3738899175007,56.3906860376445,56.3789062474693,56.363491057507,56.3448600773262,56.2106018072017,56.1834716771916,56.1896955278843,56.1964302072963,56.1816749573197,56.1596488971734,55.9313888468334,55.8499984665758,56.1223564071002,56.2378959670958,56.2229309069634,56.1683425867914,55.9700546263208,55.860279075786,55.8577728255502,55.9086685152469,55.8034057546722,55.782222744405,55.9194946251101,55.9127800368867,56.0947265559011,56.1236152558646,56.1318702656965,55.894287104257,55.8287467939951,55.8389625438857,55.8188896137501,55.8516845637132,55.8576792152289]},{"lng":[-79.6353941427214,-79.6301879833946,-79.6038207936468,-79.5922851537535,-79.6353941427214],"lat":[55.9118877739432,55.9071044851719,55.9302825852952,55.9431152253486,55.9118877739432]}],[{"lng":[-78.9243774378768,-78.9111099179268,-78.9377746578286,-78.9204101578903,-78.9243774378768],"lat":[56.111106867669,56.0997238177022,56.097499847634,56.1198730476796,56.111106867669]}],[{"lng":[-78.9199829078965,-78.9297714178587,-78.9081039379413,-78.9199829078965],"lat":[56.0844726576791,56.0964775076546,56.0712013177085,56.0844726576791]}],[{"lng":[-78.9888915975978,-78.9974517775671,-79.0702438372559,-79.0506133973356,-78.9882812475981,-78.9888915975978],"lat":[56.3119430475095,56.2786025974846,56.2216224672773,56.2857246373372,56.3281249975119,56.3119430475095]}],[{"lng":[-79.0366821274382,-79.0680389373066,-79.0752563472707,-79.0366821274382],"lat":[56.0189208973629,55.9663886972703,55.9841651872502,56.0189208973629]}],[{"lng":[-79.871108998814,-79.9033202982993,-79.8898696785059,-79.8247146495217,-79.871108998814],"lat":[59.9317436146144,59.902526854446,59.9417381245228,59.9696197448487,59.9317436146144]}],[{"lng":[-79.2247543265595,-79.1298065170469,-79.240760796471,-79.2247543265595],"lat":[55.687839506766,55.7671318070752,55.6732406567112,55.687839506766]}],[{"lng":[-78.7299804688913,-78.6965942389607,-78.7638854988193,-78.7244262689069,-78.8036956787262,-78.7788619987808,-78.7299804688913],"lat":[52.4436225880532,52.450832368126,52.4169425979769,52.4122924780654,52.4114952078841,52.4477157579425,52.4436225880532]}],[{"lng":[-79.5277786246778,-79.0477905275614,-79.06777953748,-79.0188903776823,-79.2499999965873,-79.2199096667495,-79.2421874966276,-79.413887015563,-79.3761138858179,-79.577690114261,-79.7527770925297,-79.7033309830544,-79.6688842734074,-79.7266693028034,-79.6666641134164,-79.5277786246778],"lat":[54.8594436555567,54.9403076172846,54.9183349572252,54.9252777073669,54.8758315966396,54.8922729467431,54.8919067366673,54.8233337360287,54.8452758761775,54.8230705253328,54.7733154244781,54.8011093047324,54.8024902249026,54.8063888446148,54.8372230449157,54.8594436555567]}],[{"lng":[-79.8694000088592,-79.8876876685672,-79.8940963584582,-79.8597183090053,-79.8694000088592],"lat":[59.886123654613,59.8780670145197,59.8877372644895,59.905422204665,59.886123654613]}],[{"lng":[-78.7818145787328,-78.7761077887484,-78.7938842787045,-78.7818145787328],"lat":[52.7770156879368,52.76194381795,52.7686767579086,52.7770156879368]}],[{"lng":[-79.2490234363933,-79.3244171059418,-79.3012390061057,-79.3329086259037,-79.337585445855,-79.217483516567,-79.2490234363933],"lat":[55.8155479366913,55.7403716964212,55.6739616365006,55.6645202563855,55.739929196373,55.8555030767997,55.8155479366913]}],[{"lng":[-79.9132079981234,-79.9533080974365,-79.9409179576429,-79.9123153581333,-79.9132079981234],"lat":[59.926879874401,59.9088745041881,59.9367217942598,59.9395217844084,59.926879874401]}],[{"lng":[-80.1050033445229,-80.1721114930556,-80.1472625535894,-80.0485229356543,-79.9488906775346,-79.9306259078611,-79.9570846474101,-80.1052017045478,-80.1260986140883,-80.1050033445229],"lat":[59.8127784633147,59.7662200828943,59.8248672430639,59.8823242136603,59.8630561742007,59.843982694292,59.8158721841463,59.7580909632989,59.7725219631778,59.8127784633147]}],[{"lng":[-78.8027801487257,-78.8355102486458,-78.8305358886563,-78.8027801487257],"lat":[52.4327773978864,52.4188842778076,52.4344482378198,52.4327773978864]}],[{"lng":[-79.9688873172239,-79.9254760579764,-79.9532775775047,-79.9912261868186,-79.948715197573,-79.9688873172239],"lat":[59.75944518407,59.7802810643044,59.7541313141522,59.7531700039473,59.7819099341828,59.75944518407]}],[{"lng":[-78.9415512082618,-78.9038696283772,-78.9282836883046,-78.9415512082618],"lat":[52.9819946275387,52.9830322276383,52.9730834975741,52.9819946275387]}],[{"lng":[-78.9783782980879,-78.9677810681255,-79.0023803680053,-78.9783782980879],"lat":[53.3177146874419,53.307777397471,53.3112144473746,53.3177146874419]}],[{"lng":[-78.9926757780222,-79.0000228879985,-79.0372238178612,-78.9926757780222],"lat":[53.4177246074035,53.4033851573826,53.4099998472758,53.4177246074035]}],[{"lng":[-79.0833129875946,-79.0638885476757,-79.0988769475301,-79.0833129875946],"lat":[53.8850707971478,53.877223967206,53.8796997071002,53.8850707971478]}],[{"lng":[-79.1155929574134,-79.0966644274979,-79.1322250373409,-79.1155929574134],"lat":[54.1169624270545,54.1066665571126,54.1063880870021,54.1169624270545]}],[{"lng":[-79.0322189278878,-78.9988403280122,-79.0330505378881,-79.0322189278878],"lat":[53.3647232072897,53.3455543473851,53.344970697287,53.3647232072897]}],[{"lng":[-78.9499969482405,-78.9522247282367,-79.009315488049,-78.9499969482405],"lat":[52.9461097675156,52.9249992375094,52.9053001373503,52.9461097675156]}],[{"lng":[-79.5108108442517,-79.4922637844449,-79.5144424342544,-79.5450286839688,-79.5744018537091,-79.5972289934747,-79.5565795838594,-79.575531003676,-79.5555419838618,-79.5965270934317,-79.568908683705,-79.5596923738027,-79.543891903947,-79.5108108442517],"lat":[56.769870755777,56.6586723258429,56.6472206057501,56.6762084956237,56.619506835491,56.6561279253938,56.6817016555747,56.6911849954932,56.7061500455816,56.8133506754133,56.8041839555337,56.7722778255703,56.7852783156394,56.769870755777]}],[{"lng":[-79.1292648273909,-79.1011276175158,-79.1115417474726,-79.1292648273909],"lat":[53.9151000970064,53.9049301070938,53.8961753770614,53.9151000970064]}],[{"lng":[-79.1244964574196,-79.0998611475275,-79.1514892572986,-79.1244964574196],"lat":[53.8761482170205,53.870822907097,53.8718872069346,53.8761482170205]}],[{"lng":[-79.0327758779413,-78.9938888480815,-79.0550003078595,-79.0327758779413],"lat":[53.043888087284,53.0305557273952,53.0394439672185,53.043888087284]}],[{"lng":[-79.565391533871,-79.5421752840702,-79.4818115145929,-79.48278808459,-79.4938964744939,-79.530807494177,-79.508300774378,-79.4894332845367,-79.5516662540226,-79.6387939431862,-79.565391533871],"lat":[56.3734130855067,56.424499505612,56.4452781658665,56.4260864258608,56.4331054658158,56.4113197256593,56.4039306557535,56.4158973658327,56.2949981655592,56.2580604551647,56.3734130855067]}],[{"lng":[-79.4966659444526,-79.4955520544678,-79.5433349540423,-79.4966659444526],"lat":[56.4952812158099,56.4755554158127,56.4822235056124,56.4952812158099]}],[{"lng":[-79.1243896474711,-79.0933608976049,-79.098854057584,-79.1383056574113,-79.1243896474711],"lat":[53.610557557015,53.6010513271112,53.5902976970941,53.6038818369708,53.610557557015]}],[{"lng":[-79.1112594575058,-79.1118545475051,-79.1279983474337,-79.1112594575058],"lat":[53.7264022770585,53.7162437370564,53.7221717770059,53.7264022770585]}],[{"lng":[-79.1843872071006,-79.1650009171953,-79.2766647266181,-79.1843872071006],"lat":[54.0786743168328,54.0727767868962,54.0758323665156,54.0786743168328]}],[{"lng":[-79.6614837529101,-79.5974883935464,-79.5413742040432,-79.526306144188,-79.6580429029627,-80.0114898582801,-80.0417327777648,-80.047721847668,-80.0799331570823,-80.0999374267174,-80.0904998668635,-80.0599441374103,-80.0538253675303,-80.0460357576171,-80.0387496777499,-79.980552658705,-79.915405259725,-79.926696769551,-79.8513488706304,-79.8643112104254,-79.7298660221533,-79.6614837529101],"lat":[56.3674621550694,56.4247970553695,56.5395164456262,56.5111083956876,56.322017665081,56.1788253731487,56.1825942929603,56.1704978829207,56.1925086927171,56.1882743725856,56.2343864326544,56.2435417128533,56.2232627828894,56.308559412951,56.2979507329954,56.3288879333579,56.3035278237375,56.3145141536738,56.3166503840979,56.3743705640339,56.3482589647358,56.3674621550694]}],[{"lng":[-79.1866683970841,-79.1711120571608,-79.2044448869967,-79.1866683970841],"lat":[54.104721066826,54.099166866877,54.0991668667666,54.104721066826]}],[{"lng":[-79.7963256708483,-79.7529678213907,-79.7073974519753,-79.7501907214743,-79.7611083913263,-79.8060607807335,-79.790000910943,-79.8425903202281,-79.803924550752,-79.8344421303359,-79.8138885406091,-79.7963256708483],"lat":[57.556884764556,57.6145172047812,57.5470161349921,57.4850730847765,57.5108337347263,57.5164184545002,57.5302772445842,57.5150756743099,57.542240134515,57.5319442643553,57.5605354244666,57.556884764556]}],[{"lng":[-79.1466064473457,-79.1350021373997,-79.1555786073078,-79.1466064473457],"lat":[53.7467040969473,53.7377777069841,53.7316894469181,53.7467040969473]}],[{"lng":[-78.972221368305,-78.9527740483649,-78.9405517584064,-78.9238891584542,-78.9494476283811,-78.9961090082325,-78.972221368305],"lat":[52.1119461074519,52.1102905275052,52.0791664075383,52.0844459475827,52.0719451875143,52.0924987773852,52.1119461074519]}],[{"lng":[-79.1155548075399,-79.1024551375941,-79.1105575575647,-79.1583328173556,-79.1160888675357,-79.1155548075399],"lat":[53.4513893070397,53.4562301570805,53.432498927055,53.4336128169031,53.4614257770382,53.4513893070397]}],[{"lng":[-79.0548248278579,-79.0538864078633,-79.07277678779,-79.0548248278579],"lat":[53.0520439072192,53.0422210672219,53.0475006071654,53.0520439072192]}],[{"lng":[-79.1822967471642,-79.1710205072205,-79.1943511971073,-79.1822967471642],"lat":[53.8202247568329,53.8079223568696,53.8129806467927,53.8202247568329]}],[{"lng":[-79.5273284842977,-79.5136108344193,-79.5603942840084,-79.5273284842977],"lat":[56.1090393056476,56.1027183457052,56.0811614955022,56.1090393056476]}],[{"lng":[-80.2780074905246,-80.3344115990046,-80.3265151791918,-80.2836913903349,-80.2431030113812,-80.2378234714878,-80.1666259632,-80.1633300632959,-80.2166747820444,-80.2780074905246],"lat":[59.6390685921705,59.6270751917788,59.6735191218489,59.6989135621506,59.7001609724212,59.7410240124679,59.7354736229197,59.7002792329304,59.6867065325891,59.6390685921705]}],[{"lng":[-79.6866760224146,-79.6711120525937,-79.6905517523785,-79.6866760224146],"lat":[56.9858398350189,56.9727783150907,56.9636115949978,56.9858398350189]}],[{"lng":[-79.6538085828193,-79.6526184028399,-79.7200012120492,-79.7020645022635,-79.7044525022231,-79.6538085828193],"lat":[56.8675270051593,56.8443870451622,56.9302787748518,56.916610714937,56.9519691449298,56.8675270051593]}],[{"lng":[-78.8843994086762,-78.9094467186154,-78.9066772486191,-78.8843994086762],"lat":[51.3361206076899,51.3083343476255,51.3313903776325,51.3361206076899]}],[{"lng":[-79.0335082981036,-78.9850006082654,-79.0199661281543,-79.1110229478254,-79.0335082981036],"lat":[52.1172828672777,52.1080551074164,52.0906486473171,52.0971069270435,52.1172828672777]}],[{"lng":[-79.2655029266754,-79.2483367867694,-79.2916870065295,-79.2655029266754],"lat":[54.095886226556,54.0936126666162,54.0908813464621,54.095886226556]}],[{"lng":[-79.0504379280092,-79.0147094681344,-79.0564346279899,-79.0504379280092],"lat":[52.3100738472278,52.3037109373321,52.29798125721,52.3100738472278]}],[{"lng":[-79.8007965008146,-79.8286361604708,-79.857719410027,-79.8352966203474,-79.8356933503363,-79.8158569206155,-79.8057022007391,-79.8007965008146],"lat":[57.4912414545235,57.4031867943664,57.479011534224,57.4759674043424,57.4890747043423,57.4787445044442,57.5144882145018,57.4912414545235]}],[{"lng":[-79.5864486637757,-79.5983352636702,-79.5916671737278,-79.6144103935096,-79.5864486637757],"lat":[56.0458450253837,56.020000455328,56.0400009153598,56.0305709752558,56.0458450253837]}],[{"lng":[-79.1566467275055,-79.1777801474109,-79.1422195375643,-79.1566467275055],"lat":[52.7454948368998,52.7538871768305,52.7597236569465,52.7454948368998]}],[{"lng":[-79.813278190908,-79.8381729005513,-79.8444442604715,-79.9020614496495,-79.924285879305,-79.8987274096839,-79.9172515794024,-79.8826675299148,-79.8878097398248,-79.8343810905964,-79.8248290907099,-79.7697753814489,-79.7398071218559,-79.7648391615591,-79.8067016510156,-79.7965087811437,-79.8239135607816,-79.813278190908],"lat":[56.8572845443704,56.9040374742449,56.8881874042091,56.8734817438908,56.8904266337676,56.9059791539141,56.9116668638107,56.9178886340051,56.9500007539813,56.9200477542673,56.9591064443232,56.9118156346019,56.8154907147403,56.7895927346116,56.8080558743984,56.8204956044531,56.8197021443094,56.8572845443704]}],[{"lng":[-79.4638900751951,-79.4338912954184,-79.5100021348361,-79.4638900751951],"lat":[54.8274993858262,54.8266677859488,54.8191680856313,54.8274993858262]}],[{"lng":[-79.1586074775321,-79.1331634476394,-79.1438903775976,-79.1699829074854,-79.1586074775321],"lat":[52.5790176368924,52.5778045669743,52.5641670169399,52.5675048768551,52.5790176368924]}],[{"lng":[-79.8687133699164,-79.8496780301941,-79.8665466199548,-79.8687133699164],"lat":[57.3671264541477,57.3588752642486,57.3515930141571,57.3671264541477]}],[{"lng":[-79.161315917524,-79.1433792076029,-79.1661529475139,-79.1821899374342,-79.161315917524],"lat":[52.5626831068835,52.5491027769415,52.5120162968674,52.5560913068147,52.5626831068835]}],[{"lng":[-80.2466659314768,-80.2694473109264,-80.2819824005799,-80.2656860209895,-80.2466659314768],"lat":[59.412776942313,59.3708343421478,59.4028091320721,59.420898432188,59.412776942313]}],[{"lng":[-80.2974777000505,-80.2814864904776,-80.3399963189067,-80.2974777000505],"lat":[59.5779571420194,59.5731353721268,59.5455551017134,59.5779571420194]}],[{"lng":[-79.7127990623923,-79.6997451725426,-79.762512201821,-79.7127990623923],"lat":[56.2372589048085,56.2286415048716,56.2067260645555,56.2372589048085]}],[{"lng":[-79.0580978381044,-79.0579605081072,-79.0897521979956,-79.0580978381044],"lat":[51.6587448072073,51.6465873672078,51.6441955571117,51.6587448072073]}],[{"lng":[-79.4860839750488,-79.5105590748577,-79.4783325151058,-79.4860839750488],"lat":[54.7447395257291,54.7350006056243,54.7550010657622,54.7447395257291]}],[{"lng":[-80.3078155298885,-80.2905883603626,-80.3178100396416,-80.3078155298885],"lat":[59.4113197218968,59.3901100120093,59.3775024318168,59.4113197218968]}],[{"lng":[-79.2443237270797,-79.221412657192,-79.2363738971228,-79.2552642770277,-79.2443237270797],"lat":[52.8345451366047,52.8311424266841,52.8175926166323,52.8235778765661,52.8345451366047]}],[{"lng":[-79.901214589722,-79.9105834895907,-79.9700012086468,-79.9534377988991,-79.9762115385282,-79.9238891493175,-79.8889922998469,-79.8666686901652,-79.8573226803109,-79.8284835707088,-79.8251037507649,-79.895690909787,-79.901214589722],"lat":[56.7463073638775,56.7286987238221,56.7649993834834,56.7909812835846,56.7990341134517,56.8772239637679,56.8645896839625,56.8745117140869,56.8445930441336,56.8454475342886,56.8212585343033,56.7818145739135,56.7463073638775]}],[{"lng":[-79.0993804879257,-79.1159896878656,-79.1344451877914,-79.0993804879257],"lat":[51.8173103270808,51.8059425370292,51.8197212169707,51.8173103270808]}],[{"lng":[-80.3757095077505,-80.3908080873298,-80.4119033566834,-80.424713106316,-80.4561156953253,-80.4382476558581,-80.3757095077505],"lat":[59.7180442715155,59.6807250913937,59.6881599312417,59.651504511134,59.6436767508947,59.6914749010468,59.7180442715155]}],[{"lng":[-79.447776785502,-79.4255523656614,-79.49999999512,-79.4910812351831,-79.447776785502],"lat":[54.1835021958627,54.174999235953,54.1616668656424,54.1786766056812,54.1835021958627]}],[{"lng":[-79.1949996875018,-79.2066039974541,-79.2427749572889,-79.2025833074647,-79.1949996875018],"lat":[51.9999999967716,51.9860839767324,51.9897232066077,52.0174102767459,51.9999999967716]}],[{"lng":[-79.5772247242973,-79.6061096140431,-79.6055297840435,-79.5772247242973],"lat":[54.7280540453293,54.7113876251957,54.7255248951993,54.7280540453293]}],[{"lng":[-79.5760650543527,-79.6027755641226,-79.5611114444886,-79.6222228939474,-79.6077880840703,-79.5760650543527],"lat":[54.5950469953271,54.5722236552033,54.5763893053936,54.5627784651118,54.591125485181,54.5950469953271]}],[{"lng":[-79.218505857451,-79.186058037593,-79.2936630171058,-79.2713317872097,-79.218505857451],"lat":[51.773498536693,51.7666892968028,51.7450294464273,51.764110566508,51.773498536693]}],[{"lng":[-80.4817886146169,-80.443328835869,-80.4891967544564,-80.4817886146169],"lat":[59.4882698006412,59.4819335809349,59.3900756705481,59.4882698006412]}],[{"lng":[-79.6311111437886,-79.6555557235542,-79.6193847638965,-79.6311111437886],"lat":[54.769721975082,54.7641677849648,54.7775268551377,54.769721975082]}],[{"lng":[-79.3814086865429,-79.341667176762,-79.2438888472667,-79.3255538868691,-79.3683319066336,-79.3322219768345,-79.3227767869006,-79.3782958966072,-79.4672241160661,-79.5671997053774,-79.6222228949619,-79.6516647246918,-79.6343994048221,-79.6050033550621,-79.5444412155136,-79.4823684659275,-79.3814086865429],"lat":[52.0858764560927,52.0974998462464,52.0597228966036,52.0219459463075,52.0294456461441,52.0186119062824,51.9594459463182,51.9259033161057,51.935554495744,51.9266967753049,51.9055557250481,51.9777793849053,52.0083465549889,51.997093195129,52.0094451854075,52.0556449856791,52.0858764560927]}],[{"lng":[-80.5332183526711,-80.5014877037815,-80.5707092013642,-80.5962753003833,-80.5332183526711],"lat":[59.6941223003114,59.6861610305597,59.6423683099873,59.6658668397848,59.6941223003114]}],[{"lng":[-79.2855224570892,-79.2572250372295,-79.2352905273298,-79.2403793273114,-79.3216476369141,-79.3227691669036,-79.2855224570892],"lat":[51.9649047864555,51.954166406557,51.963928216634,51.9427413866163,51.9313354463226,51.9482994063183,51.9649047864555]}],[{"lng":[-80.5379028027398,-80.5089645137507,-80.5280456231206,-80.5498046623434,-80.5379028027398],"lat":[59.4401245001774,59.4336585904074,59.4024238502431,59.4116821200698,59.4401245001774]}],[{"lng":[-80.5758285214052,-80.5440520925737,-80.556579562142,-80.5758285214052],"lat":[59.4048271098524,59.3848724301067,59.3677940299977,59.4048271098524]}],[{"lng":[-79.4049987763527,-79.3688888465699,-79.5104980456754,-79.4876098558228,-79.4049987763527],"lat":[52.2599983159989,52.2419433561415,52.2351074155572,52.2651710456563,52.2599983159989]}],[{"lng":[-80.5649642618435,-80.5792159713755,-80.5866088610408,-80.5649642618435],"lat":[59.3627967699266,59.3075904697869,59.3690795797485,59.3627967699266]}],[{"lng":[-79.7421493427006,-79.7261123628815,-79.7527770925921,-79.7421493427006],"lat":[54.6478881745247,54.6313896146061,54.6272239644682,54.6478881745247]}],[{"lng":[-80.5951843007855,-80.5703735117147,-80.5882110310733,-80.6373672191684,-80.5951843007855],"lat":[59.3033332696508,59.292381279855,59.2761726296992,59.2980384692866,59.3033332696508]}],[{"lng":[-80.5476836927019,-80.5399780029992,-80.5917968411255,-80.5476836927019],"lat":[59.1135024899769,59.0858764500305,59.0908813395985,59.1135024899769]}],[{"lng":[-80.6309127492102,-80.6788863773212,-80.6710204776148,-80.6309127492102],"lat":[59.5009193294244,59.4541663989856,59.4829101490674,59.5009193294244]}],[{"lng":[-80.6167068200119,-80.6234206897884,-80.6426314990286,-80.6167068200119],"lat":[59.2600173894503,59.2259025493791,59.233840929215,59.2600173894503]}],[{"lng":[-80.5955810210132,-80.5754775717651,-80.614913910298,-80.5955810210132],"lat":[59.0626830995557,59.0514983997222,59.0496978693845,59.0626830995557]}],[{"lng":[-79.6810989338789,-79.6480102441863,-79.6838912938564,-79.6810989338789],"lat":[53.3122901847735,53.3060913049363,53.3030548047593,53.3122901847735]}],[{"lng":[-79.5830078050123,-79.5872192349901,-79.5444412153223,-79.6450576745224,-79.6055297848423,-79.6255569446674,-79.5830078050123],"lat":[52.5825271552324,52.5525016752127,52.5446777254079,52.5283622649376,52.554504385127,52.5794448850321,52.5825271552324]}],[{"lng":[-79.7194442634197,-79.6880264237375,-79.7127685434969,-79.7194442634197],"lat":[53.5177764845858,53.4957809347444,53.4929237346191,53.5177764845858]}],[{"lng":[-79.8977813611914,-79.867774951576,-79.8754577514885,-79.9105834910291,-79.8977813611914],"lat":[53.9108314436231,53.9044456437968,53.8875503437517,53.9016723535472,53.9108314436231]}],[{"lng":[-79.7762451130226,-79.9233322013235,-79.9149780213856,-79.8727798418923,-79.8099975526225,-79.8034667927069,-79.8277740424426,-79.7762451130226],"lat":[53.0975303542752,53.066944113438,53.1464843734902,53.176109303739,53.1694946240933,53.1441268841285,53.1297225939936,53.0975303542752]}],[{"lng":[-80.0705566291011,-80.0966644186441,-80.0744476189836,-80.0077743399784,-79.8983154214801,-79.9572143507346,-79.9316558711046,-80.0105590700421,-80.0861205988799,-80.0705566291011],"lat":[53.285556782511,53.3461112923371,53.3694457924883,53.3927764829256,53.3586120535958,53.3153076132396,53.2638778633937,53.2255554129015,53.2496948124049,53.285556782511]}]]],null,"qc_simple",{"crs":{"crsClass":"L.CRS.EPSG3857","code":null,"proj4def":null,"projectedBounds":null,"options":{}},"pane":"polygon","stroke":true,"color":"#333333","weight":1,"opacity":0.9,"fill":true,"fillColor":"#6666FF","fillOpacity":0.6,"smoothFactor":1,"noClip":false},"<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>1 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>GID_0 <\/b><\/td><td align='right'>CAN <\/td><\/tr><tr><td>2<\/td><td><b>NAME_0 <\/b><\/td><td align='right'>Canada <\/td><\/tr><tr class='alt'><td>3<\/td><td><b>GID_1 <\/b><\/td><td align='right'>CAN.11_1 <\/td><\/tr><tr><td>4<\/td><td><b>NAME_1 <\/b><\/td><td align='right'>Québec <\/td><\/tr><tr class='alt'><td>5<\/td><td><b>VARNAME_1 <\/b><\/td><td align='right'>Lower Canada <\/td><\/tr><tr><td>6<\/td><td><b>NL_NAME_1 <\/b><\/td><td align='right'>NA <\/td><\/tr><tr class='alt'><td>7<\/td><td><b>TYPE_1 <\/b><\/td><td align='right'>Province <\/td><\/tr><tr><td>8<\/td><td><b>ENGTYPE_1 <\/b><\/td><td align='right'>Province <\/td><\/tr><tr class='alt'><td>9<\/td><td><b>CC_1 <\/b><\/td><td align='right'>24 <\/td><\/tr><tr><td>10<\/td><td><b>HASC_1 <\/b><\/td><td align='right'>CA.QC <\/td><\/tr><tr class='alt'><td>11<\/td><td><b>geometry <\/b><\/td><td align='right'>sfc_MULTIPOLYGON <\/td><\/tr><\/table><\/div><\/body><\/html>",{"maxWidth":800,"minWidth":50,"autoPan":true,"keepInView":false,"closeButton":true,"closeOnClick":true,"className":""},"1",{"interactive":false,"permanent":false,"direction":"auto","opacity":1,"offset":[0,0],"textsize":"10px","textOnly":false,"className":"","sticky":true},{"stroke":true,"weight":2,"opacity":0.9,"fillOpacity":0.84,"bringToFront":false,"sendToBack":false}]},{"method":"addScaleBar","args":[{"maxWidth":100,"metric":true,"imperial":true,"updateWhenIdle":true,"position":"bottomleft"}]},{"method":"addHomeButton","args":[-80.6788863773212,44.99119568,-57.1078383830923,63.697917939791,"Zoom to qc_simple","<strong> qc_simple <\/strong>","bottomright"]},{"method":"addLayersControl","args":[["CartoDB.Positron","CartoDB.DarkMatter","OpenStreetMap","Esri.WorldImagery","OpenTopoMap"],"qc_simple",{"collapsed":true,"autoZIndex":true,"position":"topleft"}]},{"method":"addLegend","args":[{"colors":["#6666FF"],"labels":["qc_simple"],"na_color":null,"na_label":"NA","opacity":1,"position":"topright","type":"factor","title":"qc_simple","extra":null,"layerId":null,"className":"info legend","group":"qc_simple"}]}],"limits":{"lat":[44.99119568,63.697917939791],"lng":[-80.6788863773212,-57.1078383830923]}},"evals":[],"jsHooks":{"render":[{"code":"function(el, x, data) {\n return (\n function(el, x, data) {\n // get the leaflet map\n var map = this; //HTMLWidgets.find('#' + el.id);\n // we need a new div element because we have to handle\n // the mouseover output separately\n // debugger;\n function addElement () {\n // generate new div Element\n var newDiv = $(document.createElement('div'));\n // append at end of leaflet htmlwidget container\n $(el).append(newDiv);\n //provide ID and style\n newDiv.addClass('lnlt');\n newDiv.css({\n 'position': 'relative',\n 'bottomleft': '0px',\n 'background-color': 'rgba(255, 255, 255, 0.7)',\n 'box-shadow': '0 0 2px #bbb',\n 'background-clip': 'padding-box',\n 'margin': '0',\n 'padding-left': '5px',\n 'color': '#333',\n 'font': '9px/1.5 \"Helvetica Neue\", Arial, Helvetica, sans-serif',\n 'z-index': '700',\n });\n return newDiv;\n }\n\n\n // check for already existing lnlt class to not duplicate\n var lnlt = $(el).find('.lnlt');\n\n if(!lnlt.length) {\n lnlt = addElement();\n\n // grab the special div we generated in the beginning\n // and put the mousmove output there\n\n map.on('mousemove', function (e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() +\n ' | x: ' + L.CRS.EPSG3857.project(e.latlng).x.toFixed(0) +\n ' | y: ' + L.CRS.EPSG3857.project(e.latlng).y.toFixed(0) +\n ' | epsg: 3857 ' +\n ' | proj4: +proj=merc +a=6378137 +b=6378137 +lat_ts=0.0 +lon_0=0.0 +x_0=0.0 +y_0=0 +k=1.0 +units=m +nadgrids=@null +no_defs ');\n } else {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n }\n });\n\n // remove the lnlt div when mouse leaves map\n map.on('mouseout', function (e) {\n var strip = document.querySelector('.lnlt');\n strip.remove();\n });\n\n };\n\n //$(el).keypress(67, function(e) {\n map.on('preclick', function(e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n var txt = document.querySelector('.lnlt').textContent;\n console.log(txt);\n //txt.innerText.focus();\n //txt.select();\n setClipboardText('\"' + txt + '\"');\n }\n });\n\n //map.on('click', function (e) {\n // var txt = document.querySelector('.lnlt').textContent;\n // console.log(txt);\n // //txt.innerText.focus();\n // //txt.select();\n // setClipboardText(txt);\n //});\n\n function setClipboardText(text){\n var id = 'mycustom-clipboard-textarea-hidden-id';\n var existsTextarea = document.getElementById(id);\n\n if(!existsTextarea){\n console.log('Creating textarea');\n var textarea = document.createElement('textarea');\n textarea.id = id;\n // Place in top-left corner of screen regardless of scroll position.\n textarea.style.position = 'fixed';\n textarea.style.top = 0;\n textarea.style.left = 0;\n\n // Ensure it has a small width and height. Setting to 1px / 1em\n // doesn't work as this gives a negative w/h on some browsers.\n textarea.style.width = '1px';\n textarea.style.height = '1px';\n\n // We don't need padding, reducing the size if it does flash render.\n textarea.style.padding = 0;\n\n // Clean up any borders.\n textarea.style.border = 'none';\n textarea.style.outline = 'none';\n textarea.style.boxShadow = 'none';\n\n // Avoid flash of white box if rendered for any reason.\n textarea.style.background = 'transparent';\n document.querySelector('body').appendChild(textarea);\n console.log('The textarea now exists :)');\n existsTextarea = document.getElementById(id);\n }else{\n console.log('The textarea already exists :3')\n }\n\n existsTextarea.value = text;\n existsTextarea.select();\n\n try {\n var status = document.execCommand('copy');\n if(!status){\n console.error('Cannot copy text');\n }else{\n console.log('The text is now on the clipboard');\n }\n } catch (err) {\n console.log('Unable to copy.');\n }\n }\n\n\n }\n ).call(this.getMap(), el, x, data);\n}","data":null}]}}</script> --- # Interactive map ```r mapView(sample_pts, layer.name = "Species richness") ``` <div id="htmlwidget-06bf1007d35c64f05147" style="width:792px;height:432px;" class="leaflet html-widget"></div> <script type="application/json" data-for="htmlwidget-06bf1007d35c64f05147">{"x":{"options":{"minZoom":1,"maxZoom":100,"crs":{"crsClass":"L.CRS.EPSG3857","code":null,"proj4def":null,"projectedBounds":null,"options":{}},"preferCanvas":false,"bounceAtZoomLimits":false,"maxBounds":[[[-90,-370]],[[90,370]]]},"calls":[{"method":"addProviderTiles","args":["CartoDB.Positron",1,"CartoDB.Positron",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"addProviderTiles","args":["CartoDB.DarkMatter",2,"CartoDB.DarkMatter",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"addProviderTiles","args":["OpenStreetMap",3,"OpenStreetMap",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"addProviderTiles","args":["Esri.WorldImagery",4,"Esri.WorldImagery",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"addProviderTiles","args":["OpenTopoMap",5,"OpenTopoMap",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"createMapPane","args":["point",440]},{"method":"addCircleMarkers","args":[[54.2655644931332,48.879532816718,49.141818771735,50.6355712510635,54.4035965778541,59.0153250463494,55.7743366084167,60.9753005854198,56.8167023183641,57.6171668199479,58.9557969128729,52.5183972823604,57.7471346674528,48.3963216453468,56.3110514463983,48.4856713113385,60.1850936845625,54.8147171169651,58.1346889137977,61.3804563939723,49.8478166555695,47.5944992323062,54.4643910524678,54.8634323431765,49.4446922701908,60.3230804244618,45.1610103491365,60.1960864909688,60.1636450763268,52.2751802407256,50.998224449494,51.4765518942759,54.6529084122664,50.6096704055187,53.9493456928928,48.4803096325022,48.9857833444766,56.3880844658564,54.7313421912406,55.0419362065212,51.8282631256873,57.8661401328275,52.6576990239621,48.0766483672707,48.6256435326446,48.489307647835,53.4202943620033,57.510186795992,51.1681715429859,46.1109319909688,52.9191586532942,58.1923702181958,49.7946043344847,49.3583222244104,50.5433560669602,60.1504580473104,56.0604281431043,56.5280646777401,55.6622164889566,51.1128612006087,60.6524734869327,49.8029035135201,51.6935460651887,50.980571352894,49.9033244694941,53.3521142115662,52.3278423789633,48.6652059749146,56.2561429677324,61.2026748016283,47.8345387309535,45.8424837956795,49.1846976286341,45.2699788656135,56.191060833761,51.310334351404,52.1116045639257,57.6222086774188,48.4876831786351,52.5407275919341,57.7855496775231,54.7927293286373,55.6304544709626,52.6550506499532,54.9576513186494,54.4787786598927,54.8963398469925,49.6079440687385,47.3251476122719,53.6733357735022,55.3222693851601,56.4932209835263,47.096867895145,53.2519592531293,52.8151503362327,55.3465916156796,47.915890302968,58.345542817509,49.4227828091845,56.6573621917736],[-78.1681484065796,-75.8583634320291,-72.5558070395379,-79.0922448392653,-77.3135152311384,-74.2426283415712,-75.7178809714992,-74.4505871762945,-69.9583550160506,-74.8579631374972,-74.3946696380331,-71.1863219805048,-73.7550364160517,-70.3631727971557,-73.0215258275772,-65.8065869208123,-74.8455469762486,-73.5646689399515,-70.4096375283475,-75.7543595884399,-70.9768537209406,-72.6768626939856,-70.447630554684,-74.9288748914818,-73.8583270128275,-67.9934373928958,-74.1826776920764,-74.7599085830889,-73.6868881934956,-76.6651785154706,-70.9045183201369,-77.7005580014717,-70.5900378868423,-61.1400989273501,-77.4397914940074,-70.5242243835244,-72.030398070197,-66.3894807818873,-68.2830872434929,-76.2878659437039,-74.9355867269306,-76.646993964922,-71.8959730234112,-77.179118390517,-78.3614971069535,-70.3659950231974,-77.9795022115465,-75.3358795441047,-62.3899541244975,-76.1180981528868,-70.4360644295388,-77.4525147039504,-68.0318266127034,-78.0847132733804,-64.7311068561808,-74.5514528184477,-71.0302914854227,-73.7509281140752,-63.9238545856928,-75.3045962040722,-72.0499344678949,-71.6367883766058,-65.352114048173,-69.8910769897604,-71.0088162153432,-67.5579538426527,-66.8913808922937,-77.7138058049872,-78.7180811452832,-74.4127122395443,-79.198359648158,-76.6272228829179,-76.0898098799496,-72.219287612266,-66.7438376270985,-77.0975252831259,-66.6964280758008,-74.6109842476182,-66.3697557426627,-69.0817130411985,-75.9570989122357,-75.2452319837057,-76.5367927954809,-70.1794989003717,-75.5257174630693,-72.2083769535347,-77.5077267472667,-67.9555719569705,-69.4962946867763,-70.6862247400107,-76.6252219030583,-74.9031870298122,-74.1125942114558,-74.8286556991372,-70.5252586749622,-75.9468879254774,-68.863248778473,-77.5773272862952,-77.8495958979821,-75.2259303892492],6,null,"Species richness",{"crs":{"crsClass":"L.CRS.EPSG3857","code":null,"proj4def":null,"projectedBounds":null,"options":{}},"pane":"point","stroke":true,"color":"#333333","weight":2,"opacity":[0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9],"fill":true,"fillColor":["#3E4989","#32B67A","#2B768E","#26828E","#20A486","#472C7A","#47C06F","#46337F","#39578C","#6ECE58","#228D8D","#7DD250","#481C6E","#ADDC30","#472C7A","#414287","#47C06F","#306A8E","#24878E","#9CD93B","#1F9F88","#1F998A","#ADDC30","#481C6E","#472C7A","#60CA60","#306A8E","#228D8D","#53C568","#7DD250","#ADDC30","#472C7A","#228D8D","#32B67A","#39578C","#228D8D","#1F9F88","#ADDC30","#2AB07F","#9CD93B","#32B67A","#3BBB75","#355E8D","#32B67A","#60CA60","#60CA60","#7DD250","#9CD93B","#2D708E","#32648E","#32648E","#3BBB75","#BDDF26","#306A8E","#DEE318","#DEE318","#1F998A","#47C06F","#39578C","#471366","#472C7A","#39578C","#472C7A","#2B768E","#26828E","#20938C","#26828E","#32648E","#471366","#47C06F","#3E4989","#3C508B","#20A486","#2B768E","#47C06F","#2B768E","#482475","#46337F","#32648E","#414287","#20938C","#443A84","#ADDC30","#3BBB75","#26828E","#414287","#355E8D","#306A8E","#228D8D","#6ECE58","#ADDC30","#2D708E","#228D8D","#47C06F","#306A8E","#414287","#3C508B","#CEE11D","#60CA60","#2B768E"],"fillOpacity":[0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6]},null,null,["<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>1 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>14 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>2 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>35 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>3 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>22 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>4 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>24 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>5 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>32 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>6 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>8 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>7 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>8 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>10 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>9 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>16 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>10 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>42 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>11 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>12 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>43 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>13 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>6 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>14 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>47 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>15 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>9 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>16 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>12 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>17 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>18 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>19 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>19 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>26 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>20 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>45 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>21 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>31 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>22 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>29 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>23 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>46 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>24 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>6 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>25 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>8 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>26 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>40 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>27 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>19 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>28 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>29 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>39 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>30 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>43 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>31 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>47 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>32 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>9 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>33 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>34 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>35 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>35 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>16 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>36 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>37 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>30 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>38 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>47 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>39 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>34 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>40 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>45 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>41 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>35 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>42 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>37 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>43 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>17 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>44 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>36 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>45 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>40 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>46 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>40 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>47 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>43 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>48 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>45 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>49 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>21 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>50 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>18 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>51 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>18 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>52 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>37 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>53 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>48 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>54 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>19 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>55 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>50 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>56 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>50 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>57 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>29 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>58 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>59 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>16 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>60 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>5 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>61 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>9 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>62 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>16 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>63 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>8 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>64 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>22 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>65 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>24 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>66 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>28 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>67 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>25 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>68 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>18 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>69 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>5 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>70 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>71 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>14 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>72 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>15 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>73 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>32 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>74 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>22 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>75 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>76 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>22 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>77 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>7 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>78 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>10 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>79 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>18 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>80 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>12 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>81 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>28 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>82 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>11 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>83 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>47 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>84 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>37 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>85 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>25 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>86 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>12 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>87 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>17 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>88 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>19 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>89 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>90 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>41 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>91 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>46 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>92 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>21 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>93 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>94 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>95 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>20 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>96 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>12 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>97 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>15 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>98 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>49 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>99 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>40 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>100 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>22 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>"],{"maxWidth":800,"minWidth":50,"autoPan":true,"keepInView":false,"closeButton":true,"closeOnClick":true,"className":""},["14","35","22","24","32","8","38","10","16","42","27","43","6","47","9","12","38","19","26","45","31","29","46","6","8","40","19","27","39","43","47","9","27","35","16","27","30","47","34","45","35","37","17","36","40","40","43","45","21","18","18","37","48","19","50","50","29","38","16","5","9","16","8","22","24","28","25","18","5","38","14","15","32","22","38","22","7","10","18","12","28","11","47","37","25","12","17","19","27","41","46","21","27","38","20","12","15","49","40","22"],{"interactive":false,"permanent":false,"direction":"auto","opacity":1,"offset":[0,0],"textsize":"10px","textOnly":false,"className":"","sticky":true},null]},{"method":"addScaleBar","args":[{"maxWidth":100,"metric":true,"imperial":true,"updateWhenIdle":true,"position":"bottomleft"}]},{"method":"addHomeButton","args":[-79.198359648158,45.1610103491365,-61.1400989273501,61.3804563939723,"Zoom to Species richness","<strong> Species richness <\/strong>","bottomright"]},{"method":"addLayersControl","args":[["CartoDB.Positron","CartoDB.DarkMatter","OpenStreetMap","Esri.WorldImagery","OpenTopoMap"],"Species richness",{"collapsed":true,"autoZIndex":true,"position":"topleft"}]},{"method":"addLegend","args":[{"colors":["#440154 , #440154 0%, #472978 11.1111111111111%, #3E4A89 22.2222222222222%, #31688E 33.3333333333333%, #26838E 44.4444444444444%, #1F9E88 55.5555555555556%, #35B878 66.6666666666667%, #6DCE59 77.7777777777778%, #B4DD2C 88.8888888888889%, #FDE725 100%, #FDE725 "],"labels":["5","10","15","20","25","30","35","40","45","50"],"na_color":null,"na_label":"NA","opacity":1,"position":"topright","type":"numeric","title":"Species richness","extra":{"p_1":0,"p_n":1},"layerId":null,"className":"info legend","group":"Species richness"}]}],"limits":{"lat":[45.1610103491365,61.3804563939723],"lng":[-79.198359648158,-61.1400989273501]}},"evals":[],"jsHooks":{"render":[{"code":"function(el, x, data) {\n return (\n function(el, x, data) {\n // get the leaflet map\n var map = this; //HTMLWidgets.find('#' + el.id);\n // we need a new div element because we have to handle\n // the mouseover output separately\n // debugger;\n function addElement () {\n // generate new div Element\n var newDiv = $(document.createElement('div'));\n // append at end of leaflet htmlwidget container\n $(el).append(newDiv);\n //provide ID and style\n newDiv.addClass('lnlt');\n newDiv.css({\n 'position': 'relative',\n 'bottomleft': '0px',\n 'background-color': 'rgba(255, 255, 255, 0.7)',\n 'box-shadow': '0 0 2px #bbb',\n 'background-clip': 'padding-box',\n 'margin': '0',\n 'padding-left': '5px',\n 'color': '#333',\n 'font': '9px/1.5 \"Helvetica Neue\", Arial, Helvetica, sans-serif',\n 'z-index': '700',\n });\n return newDiv;\n }\n\n\n // check for already existing lnlt class to not duplicate\n var lnlt = $(el).find('.lnlt');\n\n if(!lnlt.length) {\n lnlt = addElement();\n\n // grab the special div we generated in the beginning\n // and put the mousmove output there\n\n map.on('mousemove', function (e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() +\n ' | x: ' + L.CRS.EPSG3857.project(e.latlng).x.toFixed(0) +\n ' | y: ' + L.CRS.EPSG3857.project(e.latlng).y.toFixed(0) +\n ' | epsg: 3857 ' +\n ' | proj4: +proj=merc +a=6378137 +b=6378137 +lat_ts=0.0 +lon_0=0.0 +x_0=0.0 +y_0=0 +k=1.0 +units=m +nadgrids=@null +no_defs ');\n } else {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n }\n });\n\n // remove the lnlt div when mouse leaves map\n map.on('mouseout', function (e) {\n var strip = document.querySelector('.lnlt');\n strip.remove();\n });\n\n };\n\n //$(el).keypress(67, function(e) {\n map.on('preclick', function(e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n var txt = document.querySelector('.lnlt').textContent;\n console.log(txt);\n //txt.innerText.focus();\n //txt.select();\n setClipboardText('\"' + txt + '\"');\n }\n });\n\n //map.on('click', function (e) {\n // var txt = document.querySelector('.lnlt').textContent;\n // console.log(txt);\n // //txt.innerText.focus();\n // //txt.select();\n // setClipboardText(txt);\n //});\n\n function setClipboardText(text){\n var id = 'mycustom-clipboard-textarea-hidden-id';\n var existsTextarea = document.getElementById(id);\n\n if(!existsTextarea){\n console.log('Creating textarea');\n var textarea = document.createElement('textarea');\n textarea.id = id;\n // Place in top-left corner of screen regardless of scroll position.\n textarea.style.position = 'fixed';\n textarea.style.top = 0;\n textarea.style.left = 0;\n\n // Ensure it has a small width and height. Setting to 1px / 1em\n // doesn't work as this gives a negative w/h on some browsers.\n textarea.style.width = '1px';\n textarea.style.height = '1px';\n\n // We don't need padding, reducing the size if it does flash render.\n textarea.style.padding = 0;\n\n // Clean up any borders.\n textarea.style.border = 'none';\n textarea.style.outline = 'none';\n textarea.style.boxShadow = 'none';\n\n // Avoid flash of white box if rendered for any reason.\n textarea.style.background = 'transparent';\n document.querySelector('body').appendChild(textarea);\n console.log('The textarea now exists :)');\n existsTextarea = document.getElementById(id);\n }else{\n console.log('The textarea already exists :3')\n }\n\n existsTextarea.value = text;\n existsTextarea.select();\n\n try {\n var status = document.execCommand('copy');\n if(!status){\n console.error('Cannot copy text');\n }else{\n console.log('The text is now on the clipboard');\n }\n } catch (err) {\n console.log('Unable to copy.');\n }\n }\n\n\n }\n ).call(this.getMap(), el, x, data);\n}","data":null}]}}</script> --- # Interactive map ```r mapView(sample_pts, col.regions = "red", cex = 'richness', legend = FALSE, map.types = "Esri.WorldImagery") ``` <div id="htmlwidget-748bef452f0abee1eb86" style="width:792px;height:432px;" class="leaflet html-widget"></div> <script type="application/json" data-for="htmlwidget-748bef452f0abee1eb86">{"x":{"options":{"minZoom":1,"maxZoom":100,"crs":{"crsClass":"L.CRS.EPSG3857","code":null,"proj4def":null,"projectedBounds":null,"options":{}},"preferCanvas":false,"bounceAtZoomLimits":false,"maxBounds":[[[-90,-370]],[[90,370]]]},"calls":[{"method":"addProviderTiles","args":["Esri.WorldImagery",1,"Esri.WorldImagery",{"errorTileUrl":"","noWrap":false,"detectRetina":false}]},{"method":"createMapPane","args":["point",440]},{"method":"addCircleMarkers","args":[[54.2655644931332,48.879532816718,49.141818771735,50.6355712510635,54.4035965778541,59.0153250463494,55.7743366084167,60.9753005854198,56.8167023183641,57.6171668199479,58.9557969128729,52.5183972823604,57.7471346674528,48.3963216453468,56.3110514463983,48.4856713113385,60.1850936845625,54.8147171169651,58.1346889137977,61.3804563939723,49.8478166555695,47.5944992323062,54.4643910524678,54.8634323431765,49.4446922701908,60.3230804244618,45.1610103491365,60.1960864909688,60.1636450763268,52.2751802407256,50.998224449494,51.4765518942759,54.6529084122664,50.6096704055187,53.9493456928928,48.4803096325022,48.9857833444766,56.3880844658564,54.7313421912406,55.0419362065212,51.8282631256873,57.8661401328275,52.6576990239621,48.0766483672707,48.6256435326446,48.489307647835,53.4202943620033,57.510186795992,51.1681715429859,46.1109319909688,52.9191586532942,58.1923702181958,49.7946043344847,49.3583222244104,50.5433560669602,60.1504580473104,56.0604281431043,56.5280646777401,55.6622164889566,51.1128612006087,60.6524734869327,49.8029035135201,51.6935460651887,50.980571352894,49.9033244694941,53.3521142115662,52.3278423789633,48.6652059749146,56.2561429677324,61.2026748016283,47.8345387309535,45.8424837956795,49.1846976286341,45.2699788656135,56.191060833761,51.310334351404,52.1116045639257,57.6222086774188,48.4876831786351,52.5407275919341,57.7855496775231,54.7927293286373,55.6304544709626,52.6550506499532,54.9576513186494,54.4787786598927,54.8963398469925,49.6079440687385,47.3251476122719,53.6733357735022,55.3222693851601,56.4932209835263,47.096867895145,53.2519592531293,52.8151503362327,55.3465916156796,47.915890302968,58.345542817509,49.4227828091845,56.6573621917736],[-78.1681484065796,-75.8583634320291,-72.5558070395379,-79.0922448392653,-77.3135152311384,-74.2426283415712,-75.7178809714992,-74.4505871762945,-69.9583550160506,-74.8579631374972,-74.3946696380331,-71.1863219805048,-73.7550364160517,-70.3631727971557,-73.0215258275772,-65.8065869208123,-74.8455469762486,-73.5646689399515,-70.4096375283475,-75.7543595884399,-70.9768537209406,-72.6768626939856,-70.447630554684,-74.9288748914818,-73.8583270128275,-67.9934373928958,-74.1826776920764,-74.7599085830889,-73.6868881934956,-76.6651785154706,-70.9045183201369,-77.7005580014717,-70.5900378868423,-61.1400989273501,-77.4397914940074,-70.5242243835244,-72.030398070197,-66.3894807818873,-68.2830872434929,-76.2878659437039,-74.9355867269306,-76.646993964922,-71.8959730234112,-77.179118390517,-78.3614971069535,-70.3659950231974,-77.9795022115465,-75.3358795441047,-62.3899541244975,-76.1180981528868,-70.4360644295388,-77.4525147039504,-68.0318266127034,-78.0847132733804,-64.7311068561808,-74.5514528184477,-71.0302914854227,-73.7509281140752,-63.9238545856928,-75.3045962040722,-72.0499344678949,-71.6367883766058,-65.352114048173,-69.8910769897604,-71.0088162153432,-67.5579538426527,-66.8913808922937,-77.7138058049872,-78.7180811452832,-74.4127122395443,-79.198359648158,-76.6272228829179,-76.0898098799496,-72.219287612266,-66.7438376270985,-77.0975252831259,-66.6964280758008,-74.6109842476182,-66.3697557426627,-69.0817130411985,-75.9570989122357,-75.2452319837057,-76.5367927954809,-70.1794989003717,-75.5257174630693,-72.2083769535347,-77.5077267472667,-67.9555719569705,-69.4962946867763,-70.6862247400107,-76.6252219030583,-74.9031870298122,-74.1125942114558,-74.8286556991372,-70.5252586749622,-75.9468879254774,-68.863248778473,-77.5773272862952,-77.8495958979821,-75.2259303892492],[5.4,11,7.53333333333333,8.06666666666667,10.2,3.8,11.8,4.33333333333333,5.93333333333333,12.8666666666667,8.86666666666667,13.1333333333333,3.26666666666667,14.2,4.06666666666667,4.86666666666667,11.8,6.73333333333333,8.6,13.6666666666667,9.93333333333333,9.4,13.9333333333333,3.26666666666667,3.8,12.3333333333333,6.73333333333333,8.86666666666667,12.0666666666667,13.1333333333333,14.2,4.06666666666667,8.86666666666667,11,5.93333333333333,8.86666666666667,9.66666666666667,14.2,10.7333333333333,13.6666666666667,11,11.5333333333333,6.2,11.2666666666667,12.3333333333333,12.3333333333333,13.1333333333333,13.6666666666667,7.26666666666667,6.46666666666667,6.46666666666667,11.5333333333333,14.4666666666667,6.73333333333333,15,15,9.4,11.8,5.93333333333333,3,4.06666666666667,5.93333333333333,3.8,7.53333333333333,8.06666666666667,9.13333333333333,8.33333333333333,6.46666666666667,3,11.8,5.4,5.66666666666667,10.2,7.53333333333333,11.8,7.53333333333333,3.53333333333333,4.33333333333333,6.46666666666667,4.86666666666667,9.13333333333333,4.6,14.2,11.5333333333333,8.33333333333333,4.86666666666667,6.2,6.73333333333333,8.86666666666667,12.6,13.9333333333333,7.26666666666667,8.86666666666667,11.8,7,4.86666666666667,5.66666666666667,14.7333333333333,12.3333333333333,7.53333333333333],null,"sample_pts",{"crs":{"crsClass":"L.CRS.EPSG3857","code":null,"proj4def":null,"projectedBounds":null,"options":{}},"pane":"point","stroke":true,"color":"#333333","weight":2,"opacity":[0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9],"fill":true,"fillColor":"#FF0000","fillOpacity":[0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6]},null,null,["<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>1 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>14 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>2 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>35 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>3 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>22 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>4 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>24 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>5 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>32 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>6 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>8 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>7 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>8 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>10 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>9 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>16 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>10 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>42 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>11 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>12 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>43 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>13 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>6 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>14 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>47 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>15 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>9 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>16 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>12 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>17 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>18 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>19 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>19 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>26 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>20 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>45 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>21 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>31 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>22 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>29 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>23 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>46 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>24 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>6 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>25 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>8 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>26 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>40 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>27 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>19 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>28 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>29 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>39 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>30 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>43 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>31 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>47 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>32 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>9 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>33 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>34 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>35 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>35 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>16 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>36 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>37 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>30 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>38 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>47 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>39 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>34 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>40 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>45 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>41 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>35 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>42 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>37 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>43 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>17 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>44 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>36 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>45 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>40 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>46 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>40 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>47 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>43 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>48 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>45 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>49 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>21 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>50 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>18 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>51 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>18 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>52 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>37 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>53 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>48 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>54 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>19 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>55 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>50 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>56 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>50 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>57 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>29 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>58 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>59 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>16 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>60 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>5 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>61 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>9 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>62 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>16 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>63 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>8 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>64 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>22 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>65 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>24 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>66 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>28 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>67 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>25 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>68 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>18 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>69 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>5 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>70 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>71 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>14 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>72 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>15 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>73 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>32 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>74 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>22 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>75 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>76 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>22 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>77 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>7 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>78 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>10 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>79 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>18 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>80 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>12 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>81 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>28 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>82 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>11 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>83 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>47 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>84 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>37 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>85 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>25 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>86 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>12 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>87 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>17 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>88 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>19 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>89 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>90 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>41 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>91 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>46 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>92 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>21 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>93 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>27 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>94 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>38 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>95 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>20 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>96 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>12 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>97 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>15 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>98 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>49 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>99 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>40 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>","<html><head><link rel=\"stylesheet\" type=\"text/css\" href=\"lib/popup/popup.css\"><\/head><body><div class=\"scrollableContainer\"><table class=\"popup scrollable\" id=\"popup\"><tr class='coord'><td><\/td><td><b>Feature ID<\/b><\/td><td align='right'>100 <\/td><\/tr><tr class='alt'><td>1<\/td><td><b>richness <\/b><\/td><td align='right'>22 <\/td><\/tr><tr><td>2<\/td><td><b>sample_pts <\/b><\/td><td align='right'>sfc_POINT <\/td><\/tr><\/table><\/div><\/body><\/html>"],{"maxWidth":800,"minWidth":50,"autoPan":true,"keepInView":false,"closeButton":true,"closeOnClick":true,"className":""},["14","35","22","24","32","8","38","10","16","42","27","43","6","47","9","12","38","19","26","45","31","29","46","6","8","40","19","27","39","43","47","9","27","35","16","27","30","47","34","45","35","37","17","36","40","40","43","45","21","18","18","37","48","19","50","50","29","38","16","5","9","16","8","22","24","28","25","18","5","38","14","15","32","22","38","22","7","10","18","12","28","11","47","37","25","12","17","19","27","41","46","21","27","38","20","12","15","49","40","22"],{"interactive":false,"permanent":false,"direction":"auto","opacity":1,"offset":[0,0],"textsize":"10px","textOnly":false,"className":"","sticky":true},null]},{"method":"addScaleBar","args":[{"maxWidth":100,"metric":true,"imperial":true,"updateWhenIdle":true,"position":"bottomleft"}]},{"method":"addHomeButton","args":[-79.198359648158,45.1610103491365,-61.1400989273501,61.3804563939723,"Zoom to sample_pts","<strong> sample_pts <\/strong>","bottomright"]},{"method":"addLayersControl","args":["Esri.WorldImagery","sample_pts",{"collapsed":true,"autoZIndex":true,"position":"topleft"}]}],"limits":{"lat":[45.1610103491365,61.3804563939723],"lng":[-79.198359648158,-61.1400989273501]}},"evals":[],"jsHooks":{"render":[{"code":"function(el, x, data) {\n return (\n function(el, x, data) {\n // get the leaflet map\n var map = this; //HTMLWidgets.find('#' + el.id);\n // we need a new div element because we have to handle\n // the mouseover output separately\n // debugger;\n function addElement () {\n // generate new div Element\n var newDiv = $(document.createElement('div'));\n // append at end of leaflet htmlwidget container\n $(el).append(newDiv);\n //provide ID and style\n newDiv.addClass('lnlt');\n newDiv.css({\n 'position': 'relative',\n 'bottomleft': '0px',\n 'background-color': 'rgba(255, 255, 255, 0.7)',\n 'box-shadow': '0 0 2px #bbb',\n 'background-clip': 'padding-box',\n 'margin': '0',\n 'padding-left': '5px',\n 'color': '#333',\n 'font': '9px/1.5 \"Helvetica Neue\", Arial, Helvetica, sans-serif',\n 'z-index': '700',\n });\n return newDiv;\n }\n\n\n // check for already existing lnlt class to not duplicate\n var lnlt = $(el).find('.lnlt');\n\n if(!lnlt.length) {\n lnlt = addElement();\n\n // grab the special div we generated in the beginning\n // and put the mousmove output there\n\n map.on('mousemove', function (e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() +\n ' | x: ' + L.CRS.EPSG3857.project(e.latlng).x.toFixed(0) +\n ' | y: ' + L.CRS.EPSG3857.project(e.latlng).y.toFixed(0) +\n ' | epsg: 3857 ' +\n ' | proj4: +proj=merc +a=6378137 +b=6378137 +lat_ts=0.0 +lon_0=0.0 +x_0=0.0 +y_0=0 +k=1.0 +units=m +nadgrids=@null +no_defs ');\n } else {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n }\n });\n\n // remove the lnlt div when mouse leaves map\n map.on('mouseout', function (e) {\n var strip = document.querySelector('.lnlt');\n strip.remove();\n });\n\n };\n\n //$(el).keypress(67, function(e) {\n map.on('preclick', function(e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n var txt = document.querySelector('.lnlt').textContent;\n console.log(txt);\n //txt.innerText.focus();\n //txt.select();\n setClipboardText('\"' + txt + '\"');\n }\n });\n\n //map.on('click', function (e) {\n // var txt = document.querySelector('.lnlt').textContent;\n // console.log(txt);\n // //txt.innerText.focus();\n // //txt.select();\n // setClipboardText(txt);\n //});\n\n function setClipboardText(text){\n var id = 'mycustom-clipboard-textarea-hidden-id';\n var existsTextarea = document.getElementById(id);\n\n if(!existsTextarea){\n console.log('Creating textarea');\n var textarea = document.createElement('textarea');\n textarea.id = id;\n // Place in top-left corner of screen regardless of scroll position.\n textarea.style.position = 'fixed';\n textarea.style.top = 0;\n textarea.style.left = 0;\n\n // Ensure it has a small width and height. Setting to 1px / 1em\n // doesn't work as this gives a negative w/h on some browsers.\n textarea.style.width = '1px';\n textarea.style.height = '1px';\n\n // We don't need padding, reducing the size if it does flash render.\n textarea.style.padding = 0;\n\n // Clean up any borders.\n textarea.style.border = 'none';\n textarea.style.outline = 'none';\n textarea.style.boxShadow = 'none';\n\n // Avoid flash of white box if rendered for any reason.\n textarea.style.background = 'transparent';\n document.querySelector('body').appendChild(textarea);\n console.log('The textarea now exists :)');\n existsTextarea = document.getElementById(id);\n }else{\n console.log('The textarea already exists :3')\n }\n\n existsTextarea.value = text;\n existsTextarea.select();\n\n try {\n var status = document.execCommand('copy');\n if(!status){\n console.error('Cannot copy text');\n }else{\n console.log('The text is now on the clipboard');\n }\n } catch (err) {\n console.log('Unable to copy.');\n }\n }\n\n\n }\n ).call(this.getMap(), el, x, data);\n}","data":null}]}}</script> --- # Great online resources .pull-left[ #### Good tutorials for spatial data in R - [Raster analysis in R](https://mgimond.github.io/megug2017/) - [Geocomputation with R](https://geocompr.robinlovelace.net/intro.html) - [Spatial data in R](https://github.com/Pakillo/R-GIS-tutorial/blob/master/R-GIS_tutorial.md) - [Document par Nicolas Casajus (fr)](https://qcbs.ca/wiki/_media/gisonr.pdf) - [r-spatial](http://r-spatial.org/) - [Tutorial on datacamp](https://www.datacamp.com/courses/spatial-analysis-in-r-with-sf-and-raster) - [R in space - Insileco](https://insileco.github.io/2018/04/14/r-in-space---a-series/) - [Geospatial analyses & maps with R](https://mhbrice.github.io/Rspatial/Rspatial_script.html) #### Get free data - [free data at country level](http://www.diva-gis.org/gdata) - [Quebec free data](http://mffp.gouv.qc.ca/le-ministere/acces-aux-donnees-gratuites/) - [find more spatial data](https://freegisdata.rtwilson.com/) - [create shapefile on line](http://geojson.io/) - EPSG: [link1](http://spatialreference.org/); [link2](http://epsg.io/) ] .pull-right[ #### Maps in R - [Introduction to visualising spatial data in R](https://cran.r-project.org/doc/contrib/intro-spatial-rl.pdf) - [Geocomputation with R](https://geocompr.robinlovelace.net/adv-map.html) - [choropleth](https://cengel.github.io/rspatial/4_Mapping.nb.html) - [leaflet](https://rstudio.github.io/leaflet/) - [Mapview](https://r-spatial.github.io/mapview/index.html) - [tmap](https://cran.r-project.org/web/packages/tmap/vignettes/tmap-nutshell.html) - [plotly](https://plot.ly/python/maps/) - [Animated maps](https://insileco.github.io/2017/07/05/animations-in-r-time-series-of-erythemal-irradiance-in-the-st.-lawrence/) #### `sf` manipulations - [sf vignette #4](https://cran.r-project.org/web/packages/sf/vignettes/sf4.html) - [Geocomputation with R](https://geocompr.robinlovelace.net/attr.html) - [Attribute manipulations](https://insileco.github.io/2018/04/09/r-in-space---attribute-manipulations/) - [Tidy spatial data in R](http://strimas.com/r/tidy-sf/) ] --- class: inverse, center, middle # Introduction to ordination: PCA+RDA ## <i class="fa fa-sitemap" aria-hidden="true"></i> --- # Ordination of multivariate data <br> .center[] --- # Ordination of multivariate data <br> .center[] --- # Ordination of multivariate data <br> .center[] --- # Unconstrained vs constrained ordinations ### Unconstrained ordination - Reveal the main patterns in multivariate data (e.g. matrix of species abundance at different sites or matrix of environmental variables at different sites) - Describe relationship among variables of one matrix. - Descriptive method: no statistical test ### Constrained/canonical ordination - Explicitly puts into relationship two matrices: one dependent matrix and one explanatory matrix. - This approach combines the techniques of ordination and multiple regression. - Explanatory method: statistical test <br> *Both types of ordination are based upon a comparison of all possible pairs of objects (or descriptors) using association measures.* --- # Unconstrained vs constrained ordinations <br> | Response data | Explanatory variables | Analysis | |---------------|------------------------|---------------------| | 1 variable | 1 variable | Simple regression | | 1 variable | m variables | Multiple regression | | p variables | - | Simple ordination | | p variables | m variables | Canonical ordination| --- # Steps to ordination 1. Explore your data: - multivariate data, species abundance or environmental variables, 1 or 2 matrices 2. Prep your data 3. Perform an ordination analysis: - Unconstrained to describe 1 matrix - Constrained to explain the species matrix by the environmental matrix 4. Interpret the output 5. Plot 6. For constrained ordination: variable selection + test of significance --- # Packages for this section ```r library(vegan) # for all multivariate methods library(ade4) # for the Doubs dataset library(scales) # for color transparency ``` --- class: inverse, center, middle # Explore data --- # Doubs River Fish Dataset .pull-left[ Verneaux (1973) dataset: - characterization of fish communities - 27 different species - 30 different sites - 11 environmental variables Load the Doubs River species data ```r data(doubs) # available in ade4 spe <- doubs$fish env <- doubs$env ``` ] .pull.right[  ] --- # Explore environmental data ```r head(env) # first 6 rows # dfs alt slo flo pH har pho nit amm oxy bdo # 1 3 934 6.176 84 79 45 1 20 0 122 27 # 2 22 932 3.434 100 80 40 2 20 10 103 19 # 3 102 914 3.638 180 83 52 5 22 5 105 35 # 4 185 854 3.497 253 80 72 10 21 0 110 13 # 5 215 849 3.178 264 81 84 38 52 20 80 62 # 6 324 846 3.497 286 79 60 20 15 0 102 53 ``` --- # Explore environmental data ```r pairs(env) ``` <img src="index_files/figure-html/unnamed-chunk-91-1.png" width="792" style="display: block; margin: auto;" /> --- # Explore species data ```r head(spe) # Cogo Satr Phph Neba Thth Teso Chna Chto Lele Lece Baba Spbi Gogo Eslu Pefl Rham Legi Scer # 1 0 3 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 # 2 0 5 4 3 0 0 0 0 0 0 0 0 0 0 0 0 0 0 # 3 0 5 5 5 0 0 0 0 0 0 0 0 0 1 0 0 0 0 # 4 0 4 5 5 0 0 0 0 0 1 0 0 1 2 2 0 0 0 # 5 0 2 3 2 0 0 0 0 5 2 0 0 2 4 4 0 0 2 # 6 0 3 4 5 0 0 0 0 1 2 0 0 1 1 1 0 0 0 # Cyca Titi Abbr Icme Acce Ruru Blbj Alal Anan # 1 0 0 0 0 0 0 0 0 0 # 2 0 0 0 0 0 0 0 0 0 # 3 0 0 0 0 0 0 0 0 0 # 4 0 1 0 0 0 0 0 0 0 # 5 0 3 0 0 0 5 0 0 0 # 6 0 2 0 0 0 1 0 0 0 ``` --- # Explore species data Take a look at the distribution of species frequencies ```r ab <- table(unlist(spe)) barplot(ab, las = 1, col = grey(5:0/5), xlab = "Abundance class", ylab = "Frequency") ``` <img src="index_files/figure-html/unnamed-chunk-93-1.png" width="576" style="display: block; margin: auto;" /> .alert[Note the proportion of 0s.] --- # Double zero **How do we interpret the double absence of species in a matrix of species abundances (or presence-absence)?** <img style="float: right; width:50%;margin: 1%" src="images/double-zero.png"> - the .red[*presence*] of a species in 2 sites indicates .red[ecological resemblance] between these 2 sites - the sites provide a set of minimal conditions allowing the species to survive. - the .red[*absence*] of a species from 2 sites may be due to a .red[variety of causes]: competition, several dimensions of the niche (pH, light, temperature...), random dispersal process. .alert[A measured 0 (e.g 0mg/L, 0°C) is not the same than a 0 representing an absence of observation] --- class: inverse, center, middle # Data preparation --- # Distance measures .comment[Ordination analysis are based on the comparison of all possible pairs of objects. PCA and RDA are based on the Euclidean distance.] <br> Here we want to measure the ecological resemblance (distance or similarity) among sites. <img style="float: right; width:30%;margin: 1%" src="images/distMes.png"> - To compare sites based on their .alert[environmental variables], we can use the .alert[Eucidean distance] - To compare sites based on their .alert[species composition], we don't want to consider double-zero as a similarity. .alert[Euclidean distance is not appropriate]. - Solution 1: **Pre-transform** the species matrix to trick the PCA or RDA to use an appropriate distance such as Hellinger. - Solution 2: Run a correspondance analysis (CA) which is based on the Chi-square distance - Solution 3: Compute a distance matrix from the species matrix and run a PCoA --- # Standardization of environmental data Standardizing environmental variables is necessary as you cannot compare the effects of variables with different units ```r env.z <- decostand(env, method = "standardize") # env.z <- scale(env) ``` Standardization centers (mean `\(\mu\)` = 0) and scales (standard deviation `\(\sigma\)` = 1) the variables ```r apply(env.z, 2, mean) # dfs alt slo flo pH har # 2.307905e-17 1.814232e-18 -1.196236e-17 4.219715e-17 -5.547501e-18 3.348595e-16 # pho nit amm oxy bdo # 2.662801e-17 -4.104360e-17 -4.357409e-17 -2.636780e-16 6.859928e-17 apply(env.z, 2, sd) # dfs alt slo flo pH har pho nit amm oxy bdo # 1 1 1 1 1 1 1 1 1 1 1 ``` --- # Transforming species community data <br> .center[ ] --- # Transforming species community data .alert[Hellinger transformation] ```r spe.hel <- decostand(spe, method = "hellinger") ``` <br> ### Other transformation methods: - `method = "chi.square"`  **Chi-2 transformation**. Using this transformation in a PCA yields similar results to that of a correspondance analysis (CA). - `method = "normalize"`  **Chord transformation** - `method = "total"`  **Species profile transformation** > According to [Legendre & Gallagher](http://adn.biol.umontreal.ca/~numericalecology/Reprints/Legendre_&_Gallagher.pdf): The Hellinger and chord transformations appear to be the best for general use with species data. --- class: inverse, center, middle # Unconstrained ordination ## PCA --- # Principal Component Analysis (PCA) - Starting from a multidimensional sites x descriptors matrix - where the descriptors can be species abundance (or presence-absence) or environmental variables - A PCA will preserve the maximum amount of variation in the data in a reduced number of dimensions (usually we use the first 2 dimensions) - The resulting, synthetic variables are orthogonal (i.e. perpendicular and therefore uncorrelated) .center[ ] --- # PCA - Multidimensional case <br/><br/> - **PC1**  axis that maximizes the variance of the points that are projected perpendicularly onto the axis. - **PC2**  must be perpendicular to PC1, but the direction is again the one in which variance is maximized when points are perpendicularly projected - **PC3**  and so on: perpendicular to the first two axes <br/> .red[For a matrix containing *p* descriptors, a PCA produces *p* axes which are ordered according to the % of variation of the data they explain] --- # PCA - Let's try it on the fish dataset! - For both PCA and RDA, we can use the `rda()` function from the vegan package - `rda(Y ~ X)` or `rda(Y, X)`  RDA - `rda(Y)` or `rda(X)`  PCA --- # PCA - Run a PCA on the Hellinger-transformed fish data and extract the results ```r spe.h.pca <- rda(spe.hel) summary(spe.h.pca) ... # # Call: # rda(X = spe.hel) # # Partitioning of variance: # Inertia Proportion # Total 0.5023 1 # Unconstrained 0.5023 1 # # Eigenvalues, and their contribution to the variance # # Importance of components: # PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 # Eigenvalue 0.2491 0.06592 0.04615 0.03723 0.02125 0.01662 0.01477 0.01297 # Proportion Explained 0.4959 0.13122 0.09188 0.07412 0.04230 0.03309 0.02940 0.02582 # Cumulative Proportion 0.4959 0.62715 0.71903 0.79315 0.83544 0.86853 0.89794 0.92376 # PC9 PC10 PC11 PC12 PC13 PC14 PC15 PC16 # Eigenvalue 0.01054 0.006666 0.00504 0.004258 0.002767 0.002612 0.001505 0.001387 # Proportion Explained 0.02098 0.013269 0.01003 0.008477 0.005507 0.005200 0.002996 0.002761 # Cumulative Proportion 0.94474 0.958011 0.96804 0.976521 0.982029 0.987229 0.990225 0.992986 ... ``` --- #PCA - Interpretation of Output ```r summary(spe.h.pca) ... # Call: # rda(X = spe.hel) # # Partitioning of variance: # Inertia Proportion # Total 0.5023 1 # Unconstrained 0.5023 1 ... ``` - `Inertia`  general term for "variation" in the data. - Total variance of the dataset (here the fish species) = 0.5023 - In PCA, note that the "Total" and "Unconstrained" portion of the inertia is identical --- # PCA - Interpretation of Output ```r summary(spe.h.pca) ... # Call: # rda(X = spe.hel) # # Partitioning of variance: # Inertia Proportion # Total 0.5023 1 # Unconstrained 0.5023 1 # # Eigenvalues, and their contribution to the variance # # Importance of components: # PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 # Eigenvalue 0.2491 0.06592 0.04615 0.03723 0.02125 0.01662 0.01477 0.01297 # Proportion Explained 0.4959 0.13122 0.09188 0.07412 0.04230 0.03309 0.02940 0.02582 # Cumulative Proportion 0.4959 0.62715 0.71903 0.79315 0.83544 0.86853 0.89794 0.92376 ... ``` - `Eigenvalue`  measures the amount of variation represented by each ordination axe (Principal Component or PC) - One eigenvalue associated to each PC (in this output there are 27 PCs, as this is the number of species) <br/> .center[**0.2491 + 0.06592 + ... = 0.5023 Total inertia**] --- # PCA - Interpretation of Output ```r summary(spe.h.pca) ... # Call: # rda(X = spe.hel) # # Partitioning of variance: # Inertia Proportion # Total 0.5023 1 # Unconstrained 0.5023 1 # # Eigenvalues, and their contribution to the variance # # Importance of components: # PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 # Eigenvalue 0.2491 0.06592 0.04615 0.03723 0.02125 0.01662 0.01477 0.01297 # Proportion Explained 0.4959 0.13122 0.09188 0.07412 0.04230 0.03309 0.02940 0.02582 # Cumulative Proportion 0.4959 0.62715 0.71903 0.79315 0.83544 0.86853 0.89794 0.92376 ... ``` - `Proportion Explained`  proportion of variation accounted for, by dividing the eigenvalues by the total inertia. <br/> .center[**0.2491/0.5023 = 0.4959 or 49.59%**] --- # PCA - Interpretation of Output ```r summary(spe.h.pca) ... # Species scores # # PC1 PC2 PC3 PC4 PC5 PC6 # Cogo 0.17113 0.08669 -0.060772 0.2536941 -0.027774 0.0129709 # Satr 0.64097 0.02193 -0.232895 -0.1429053 -0.059150 0.0013299 # Phph 0.51106 0.19774 0.165053 0.0203364 0.105582 0.1226508 # Neba 0.38002 0.22219 0.235145 -0.0344577 0.126570 0.0570437 # Thth 0.16679 0.06494 -0.087248 0.2444247 0.016349 0.0537362 # Teso 0.07644 0.14707 -0.041152 0.2304754 -0.104984 -0.0424943 # Chna -0.18392 0.05238 -0.042963 0.0222676 0.071349 0.0299974 # Chto -0.14601 0.17844 -0.029561 0.0622957 -0.002615 -0.0839282 ... ``` - `Species scores`  coordinates of all descriptors in the multidimensional space of the PCA. <br> - Species always refers to your descriptors (the columns in your matrix), here the fish species. Even if you run a PCA on environmental variables, the descriptors will still be called Species scores --- # PCA - Interpretation of Output ```r summary(spe.h.pca) ... # Site scores (weighted sums of species scores) # # PC1 PC2 PC3 PC4 PC5 PC6 # 1 0.370801 -0.49113 -1.018411 -0.58387 -0.491672 -0.622062 # 2 0.507019 -0.05688 -0.175969 -0.42418 0.407790 0.160725 # 3 0.464652 0.02937 -0.067360 -0.49566 0.324339 0.313053 # 4 0.299434 0.19037 0.241315 -0.54538 0.009838 0.197974 # 5 -0.003672 0.13483 0.515723 -0.53640 -0.796183 -0.208554 # 6 0.212943 0.16142 0.538108 -0.44366 -0.138553 -0.066196 # 7 0.440596 -0.01853 0.174782 -0.31336 0.171713 0.131172 # 8 0.032182 -0.53492 -0.354289 -0.07870 -0.125287 -0.632592 ... ``` - `Site scores`  coordinates of all sites in the multidimensional space of the PCA. <br> - Site refers to the rows in your dataset, here the different sites along the Doubs river (but it can be points in time, etc) --- # Selecting Important PCs Select PCs which capture more variance than the average (Kaiser-Guttman criterion) ```r ev <- spe.h.pca$CA$eig # extract eigenvalues barplot(ev, main = "Eigenvalues", col = "grey", las = 2) abline(h = mean(ev), col = "red3", lwd = 2) ``` <img src="index_files/figure-html/unnamed-chunk-103-1.png" width="720" style="display: block; margin: auto;" /> --- # PCA - environmental variables We can also run PCAs on standardized environmental variables, to compare environmental conditions of sites or how environmental variables are correlated... - Run a PCA on the standardized environmental variables and extract the results ```r env.pca <- rda(env.z) summary(env.pca) ... # # Call: # rda(X = env.z) # # Partitioning of variance: # Inertia Proportion # Total 11 1 # Unconstrained 11 1 # # Eigenvalues, and their contribution to the variance # # Importance of components: # PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 # Eigenvalue 6.3216 2.2316 1.00420 0.50068 0.37518 0.24797 0.16647 0.107161 # Proportion Explained 0.5747 0.2029 0.09129 0.04552 0.03411 0.02254 0.01513 0.009742 # Cumulative Proportion 0.5747 0.7776 0.86886 0.91437 0.94848 0.97102 0.98616 0.995898 # PC9 PC10 PC11 # Eigenvalue 0.02354 0.017259 0.0043137 # Proportion Explained 0.00214 0.001569 0.0003922 # Cumulative Proportion 0.99804 0.999608 1.0000000 ... ``` --- # PCA - environmental variables - Plot the eigenvalues above average ```r ev <- env.pca$CA$eig # extract eigenvalues barplot(ev, main = "Eigenvalues", col = "grey", las = 2) abline(h = mean(ev), col = "red3", lwd = 2) ``` <img src="index_files/figure-html/unnamed-chunk-105-1.png" width="720" style="display: block; margin: auto;" /> --- # PCA - Visualization The abundance of information produced by PCA is easier to understand and interpret using biplots to visualize patterns - We can produce a quick biplot of the PCA using the function `biplot()` in base R ```r biplot(spe.h.pca) ``` <img src="index_files/figure-html/unnamed-chunk-106-1.png" width="432" style="display: block; margin: auto;" /> --- # PCA - Scaling ```r biplot(spe.h.pca, scaling = 1, main = "Scaling 1 = distance biplot") biplot(spe.h.pca, scaling = 2, main = "Scaling 2 = correlation biplot") ``` <img src="index_files/figure-html/unnamed-chunk-107-1.png" width="720" style="display: block; margin: auto;" /> - In a PCA, we interpret the .purple[**correlation among species**] by looking at the angles among their arrows, while we interpret the .purple[**distance among sites**]. - However, we cannot optimally display sites and species together in a PCA biplot. --- # PCA - Scaling <img src="index_files/figure-html/unnamed-chunk-108-1.png" width="648" style="display: block; margin: auto;" /> .pull-left[ **Scaling 1**: - distances among objects are approximations of Euclidean distances; - the angles among descriptors vector are meaningless. .alert[Best for interpreting relationships among objects (sites)!] ] .pull-right[**Scaling 2** (Default): - angles between descriptors vectors reflect their correlations; - distances among objects are not approximations of Euclidean distances. .alert[Best for interpreting relationships among descriptors (species)!] ] --- # Customized biplot <br> - Extract species and site scores along the 1st and 2nd PC: ```r spe.scores <- scores(spe.h.pca, display = "species", choices = 1:2) site.scores <- scores(spe.h.pca, display = "sites", choices = 1:2) ``` - Extract % of variation on the 1st and 2nd PC: ```r prop1 <- round(spe.h.pca$CA$eig[1]/sum(spe.h.pca$CA$eig)*100, 2) prop2 <- round(spe.h.pca$CA$eig[2]/sum(spe.h.pca$CA$eig)*100, 2) ``` --- # Customized biplot Empty plot with informative axis labels ```r plot(spe.h.pca, type = "none", xlab = paste0("PC1 (", prop1, "%)"), ylab = paste0("PC2 (", prop2, "%)")) ``` <img src="index_files/figure-html/unnamed-chunk-111-1.png" width="432" style="display: block; margin: auto;" /> --- # Customized biplot Add points for sites ```r points(site.scores, pch = 21, bg = "steelblue", cex = 1.2) ``` <img src="index_files/figure-html/unnamed-chunk-112-1.png" width="432" style="display: block; margin: auto;" /> --- # Customized biplot Add arrows for species ```r arrows(x0 = 0, y0 = 0, x1 = spe.scores[,1], y1 = spe.scores[,2], length = 0.1) # size of arrow head ``` <img src="index_files/figure-html/unnamed-chunk-113-1.png" width="432" style="display: block; margin: auto;" /> --- # Customized biplot Add labels for species ```r text(spe.scores[,1], spe.scores[,2], labels = rownames(spe.scores), col = "red3", cex = 0.8) ``` <img src="index_files/figure-html/unnamed-chunk-114-1.png" width="432" style="display: block; margin: auto;" /> --- # Customized biplot Complete code for the biplot ```r plot(spe.h.pca, type = "none", xlab = paste0("PC1 (", prop1, "%)"), ylab = paste0("PC2 (", prop2, "%)")) points(site.scores, pch = 21, bg = "steelblue", cex = 1.2) arrows(x0 = 0, y0 = 0, x1 = spe.scores[,1], y1 = spe.scores[,2], length = 0.1) text(spe.scores[,1], spe.scores[,2], labels = rownames(spe.scores), col = "red3", cex = 0.8) ``` --- class: inverse, center, middle # Constrained ordinations ## Redundancy analysis - RDA --- # RDA - RDA = combination of multiple regressions and a PCA. - It allows us to identify and test relationships between a response matrix and explanatory matrix or matrices .center[ ] - Explanatory variables can be quantitative, qualitative, or binary (0/1). <br> - .alert[Transform] species data and .alert[standardize] quantitative variables prior to running a RDA. --- # Run the RDA - Model the effect of all environmental variables on fish community composition ```r spe.rda <- rda(spe.hel ~., data = env.z) # "~." indicates all variables # or spe.rda <- rda(X = spe.hel, Y = env.z) ``` - .comment[It is better to use the formula interface which is more flexible (allows for factors and interaction among variables)], for example: - `rda(spe.hel ~ var1 + var2*var3 + fac, data = env.z)` - You can also control for a group of variables, for example you are interested in the effect var1,2,3 while holding constant var4,5: - `rda(spe.hel ~ var1 + var2*var3 + Condition(var4 + var5), data = env.z)` --- # RDA output in R ```r summary(spe.rda) ... # # Call: # rda(formula = spe.hel ~ dfs + alt + slo + flo + pH + har + pho + nit + amm + oxy + bdo, data = env.z) # # Partitioning of variance: # Inertia Proportion # Total 0.5023 1.0000 # Constrained 0.3557 0.7081 # Unconstrained 0.1467 0.2919 ... ``` - **`Constrained Proportion`**  variance of the response matrix explained by the environmental matrix - .alert[(0.3557/0.5023 = 70.81%)] - Equivalent of unadjusted R2 - **`Unconstained Proportion`**  unexplained variance in the response matrix (residuals) - .alert[(0.1467/0.5023 = 29.21%)] --- # RDA output in R ```r summary(spe.rda) ... # Eigenvalues, and their contribution to the variance # # Importance of components: # RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 # Eigenvalue 0.2218 0.0545 0.03245 0.02186 0.009033 0.006009 0.003783 0.002822 # Proportion Explained 0.4415 0.1085 0.06460 0.04351 0.017983 0.011962 0.007532 0.005618 # Cumulative Proportion 0.4415 0.5500 0.61463 0.65814 0.676119 0.688082 0.695614 0.701231 # RDA9 RDA10 RDA11 PC1 PC2 PC3 PC4 PC5 # Eigenvalue 0.001944 0.0008515 0.0006343 0.04218 0.02887 0.01910 0.01363 0.01060 # Proportion Explained 0.003869 0.0016951 0.0012628 0.08396 0.05746 0.03801 0.02713 0.02111 # Cumulative Proportion 0.705100 0.7067955 0.7080583 0.79202 0.84948 0.88749 0.91462 0.93573 # PC6 PC7 PC8 PC9 PC10 PC11 PC12 # Eigenvalue 0.008346 0.00732 0.004500 0.004236 0.002861 0.001931 0.001015 # Proportion Explained 0.016615 0.01457 0.008958 0.008432 0.005696 0.003845 0.002020 # Cumulative Proportion 0.952346 0.96692 0.975876 0.984308 0.990004 0.993848 0.995868 ... ``` - **`Eigenvalues, and their contribution to the variance`**  eigenvalues for the canonical axes (RDA1 to RDA11) and the unconstrained axes (PC1 to PC18), as well as the cumulative proportion of .purple[**variance explained by the RDA axes**] and .purple[**represented by the residual PC axes**]. - The first 2 RDA axes cumulatively explain 55% of the fish community variance. --- # Selecting variables Using .alert[stepwise selection], we can select the explanatory variables that are significant. ```r # Forward selection of variables: mod0 <- rda(spe.hel ~ 1, data = env.z) # Model with intercept only rda.sel <- ordiR2step(mod0, # lower model limit scope = formula(spe.rda), # upper model limit (full model) direction = "forward", # could be backward or both R2scope = TRUE, # can't surpass the full model's R2 step = 1000, trace = FALSE) # change to TRUE to see the selection process! ``` .comment[Here, we are essentially adding one variable at a time, and retaining it if it significantly increases the model's adjusted R2.] --- # Selecting variables - Which variables are retained by the forward selection? ```r rda.sel$call # rda(formula = spe.hel ~ dfs + oxy + bdo, data = env.z) ``` - What is the **adjusted R2** of the RDA with the selected variables? ```r RsquareAdj(rda.sel) # $r.squared # [1] 0.5645572 # # $adj.r.squared # [1] 0.5143137 ``` How would you report these results in a paper? - .comment[The selected environmental variables (dfs + oxy + bdo) explain .alert[51.43%] of the variation in fish community composition in the 30 sites along the Doubs river.] - Note: you should report the adjusted R2 which takes into account the number of explanatory variables. --- # Significance testing Use `anova.cca()` to test the significance of your RDA. ```r anova.cca(rda.sel, step = 1000) # Permutation test for rda under reduced model # Permutation: free # Number of permutations: 999 # # Model: rda(formula = spe.hel ~ dfs + oxy + bdo, data = env.z) # Df Variance F Pr(>F) # Model 3 0.28360 11.236 0.001 *** # Residual 26 0.21874 # --- # Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` You can also test the significance of each axis! ```r anova.cca(rda.sel, step = 1000, by = "axis") ``` ... and of each variable! ```r anova.cca(rda.sel, step = 1000, by = "term") ``` .center[ .alert[ If your global test is not significant, RDA results should not be interpreted!] ] --- # RDA - Visualization RDA allows a **simultaneous visualization** of your response and explanatory variables ```r ordiplot(rda.sel, scaling = 1, type = "text") ordiplot(rda.sel, scaling = 2, type = "text") ``` <img src="index_files/figure-html/unnamed-chunk-125-1.png" width="792" style="display: block; margin: auto;" /> .pull-left[ **Scaling 1** distances among objects reflect their similarities ] .pull-right[ **Scaling 2** angles between variables (species and explanatory variables) reflect their correlation ] --- # RDA triplot - Scaling ### Scaling 1 – .comment[distance triplot] .pull-left[ <img src="index_files/figure-html/unnamed-chunk-126-1.png" width="432" style="display: block; margin: auto;" /> ] .pull-right[ - .alert[Distances among sites] approximate their Euclidean distances, i.e. sites that are close together share similar species. Idem for centroids of qualitative variables. - .alert[Projecting a site at right angle] on a response variable or a quantitative explanatory variable approximates the value of the site along that variable. - The .alert[angles between response and explanatory variables] reflect their correlations (*but not the angles among response variables*). ] --- # RDA triplot - Scaling ### Scaling 2 - .comment[correlation triplot] .pull-left[ <img src="index_files/figure-html/unnamed-chunk-127-1.png" width="432" style="display: block; margin: auto;" /> ] .pull-right[ - The .alert[angles between *all* vector variables] reflect their correlations. - Distances among sites do **not** approximate their Euclidean distances. Idem for centroids of qualitative variables. - .alert[Projecting a site at right angle] on a response variable or a quantitative explanatory variable approximates the value of the site along that variable. - Environmental arrows that are .alert[along the first axis and longer] are more important to explain the variation in the community matrix. ] --- # Customizing RDA triplot Both `plot()` and `ordiplot()` make quick and simple ordination plots, but you can customize your plots by manually setting the aesthetics of points, text, and arrows. - Extract species and site scores along the 1st and 2nd RDA: ```r spe.scores <- scores(rda.sel, display = "species", choices = 1:2) site.scores <- scores(rda.sel, display = "sites", choices = 1:2) env.scores <- scores(rda.sel, display = "bp", choices= 1:2) # default: scaling = 2 ``` - Extract % of variation on the 1st and 2nd RDA: ```r prop1 <- round(rda.sel$CCA$eig[1]/sum(rda.sel$CCA$eig)*100, 2) prop2 <- round(rda.sel$CCA$eig[2]/sum(rda.sel$CCA$eig)*100, 2) ``` --- # Customizing RDA triplot ```r # Empty plot with axes plot(rda.sel, type = "none", xlab = paste0("RDA1 (", prop1, "%)"), ylab = paste0("RDA2 (", prop2, "%)")) # Points for sites points(site.scores, pch = 19, col = alpha("grey", 0.5), cex = 1.1) # Arrows and text for species arrows(x0 = 0, y0 = 0, x1 = spe.scores[,1], y1 = spe.scores[,2], length = 0.1, col = "grey15") text(spe.scores[,1], spe.scores[,2]+sign(spe.scores[,2])*.02, # shift labels labels = rownames(spe.scores), col = "grey15", cex = 0.9) # Arrows and text for environmental variables arrows(x0 = 0, y0 = 0, x1 = env.scores[,1], y1 = env.scores[,2], length = 0.1, lwd = 1.5, col = "#CC3311") text(env.scores[,1]+sign(env.scores[,1])*.05, # shift labels env.scores[,2], labels = rownames(env.scores), col = "#CC3311", cex = 1, font = 2) ``` --- # Customizing RDA triplot <img src="index_files/figure-html/unnamed-chunk-131-1.png" width="504" style="display: block; margin: auto;" /> --- # Customizing RDA triplot ```r spe.scores <- spe.scores * 1.4 ``` <img src="index_files/figure-html/unnamed-chunk-132-1.png" width="504" style="display: block; margin: auto;" /> --- # Great online resources - Multivariate analyses - dissimilarity measures, clustering, PCA, CA...: - [en](https://wiki.qcbs.ca/r_workshop9) - [fr](https://wiki.qcbs.ca/r_atelier9) - Advance multivariate analyses - RDA, CCA, MRT: - [en](https://wiki.qcbs.ca/r_workshop10) - [fr](https://wiki.qcbs.ca/r_atelier10) - Book with R scripts - [Numerical Ecology with R](http://adn.biol.umontreal.ca/~numericalecology/numecolR/) - [First edition available online](http://jinliangliu.weebly.com/uploads/2/5/7/8/25781074/numerical_ecology_with_r.pdf) --- class: inverse, center, middle # Thank you for attending this workshop!